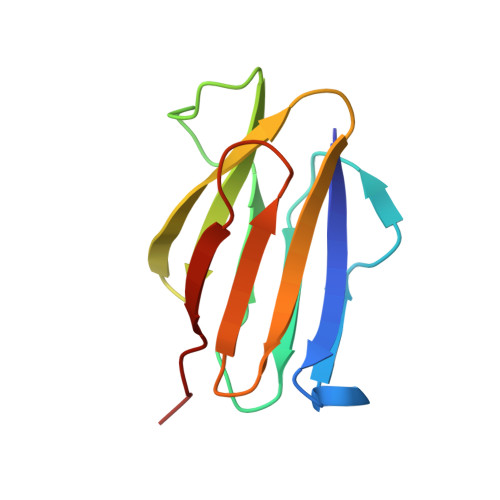
The Structure of Physarum polycephalum hemagglutinin I suggests a minimal carbohydrate recognition domain of legume lectin fold
Kouno, T., Watanabe, N., Sakai, N., Nakamura, T., Nabeshima, Y., Morita, M., Mizuguchi, M., Aizawa, T., Demura, M., Imanaka, T., Tanaka, I., Kawano, K.(2011) J Mol Biol 405: 560-569
- PubMed: 21094650
- DOI: https://doi.org/10.1016/j.jmb.2010.11.024
- Primary Citation of Related Structures:
3A5P - PubMed Abstract:
Physarum polycephalum hemagglutinin I (HA1) is a 104-residue protein that is secreted to extracellular space. The crystal structure of HA1 has a β-sandwich fold found among lectin structures, such as legume lectins and galectins. Interestingly, the β-sandwich of HA1 lacks a jelly roll motif and is essentially composed of two simple up-and-down β-sheets. This up-and-down β-sheet motif is well conserved in other legume lectin-like proteins derived from animals, plants, bacteria, and viruses. It is more noteworthy that the up-and-down β-sheet motif includes many residues that make contact with the target carbohydrates. Our NMR data demonstrate that HA1 lacking a jelly roll motif also binds to its target glycopeptide. Taken together, these data show that the up-and-down β-sheet motif provides a fundamental scaffold for the binding of legume lectin-like proteins to the target carbohydrates, and the structure of HA1 suggests a minimal carbohydrate recognition domain.
Organizational Affiliation:
Department of Structural Biology, University of Toyama, Toyama 930-0194, Japan. konox005@umn.edu