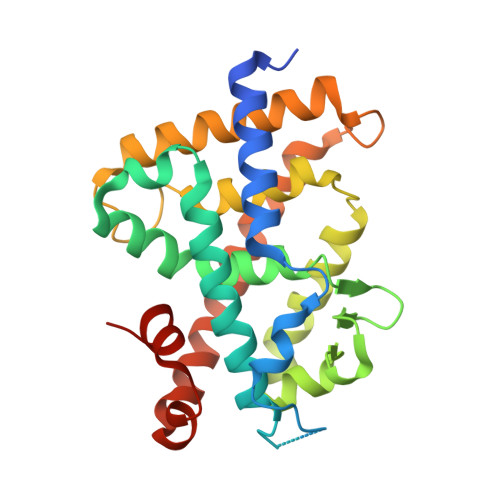
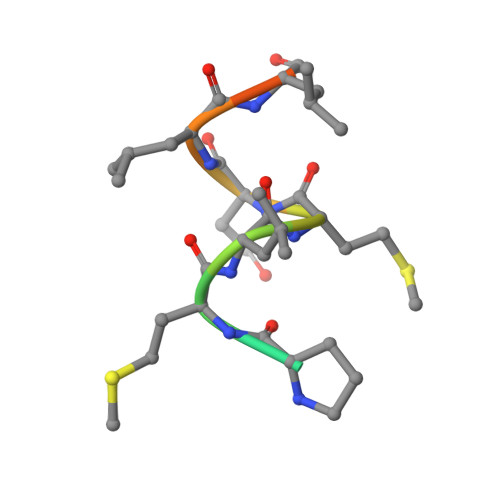
Structural basis of the histidine-mediated vitamin D receptor agonistic and antagonistic mechanisms of (23S)-25-dehydro-1alpha-hydroxyvitamin D(3)-26,23-lactone
Kakuda, S., Ishizuka, S., Eguchi, H., Mizwicki, M.T., Norman, A.W., Takimoto-Kamimura, M.(2010) Acta Crystallogr D Biol Crystallogr 66: 918-926
- PubMed: 20693691
- DOI: https://doi.org/10.1107/S0907444910020810
- Primary Citation of Related Structures:
3A2H, 3A2I, 3A2J - PubMed Abstract:
TEI-9647 antagonizes vitamin D receptor (VDR) mediated genomic actions of 1alpha,25(OH)2D3 in human cells but is agonistic in rodent cells. The presence of Cys403, Cys410 or of both residues in the C-terminal region of human VDR (hVDR) results in antagonistic action of this compound. In the complexes of TEI-9647 with wild-type hVDR (hVDRwt) and H397F hVDR, TEI-9647 functions as an antagonist and forms a covalent adduct with hVDR according to MALDI-TOF MS. The crystal structures of complexes of TEI-9647 with rat VDR (rVDR), H305F hVDR and H305F/H397F hVDR showed that the agonistic activity of TEI-9647 is caused by a hydrogen-bond interaction with His397 or Phe397 located in helix 11. Both biological activity assays and the crystal structure of H305F hVDR complexed with TEI-9647 showed that the interaction between His305 and TEI-9647 is crucial for antagonist activity. This study indicates the following stepwise mechanism for TEI-9647 antagonism. Firstly, TEI-9647 forms hydrogen bonds to His305, which promote conformational changes in hVDR and draw Cys403 or Cys410 towards the ligand. This is followed by the formation of a 1,4-Michael addition adduct between the thiol (-SH) group of Cys403 or Cys410 and the exo-methylene group of TEI-9647.
Organizational Affiliation:
Teijin Institute for Bio-Medical Research, Japan.