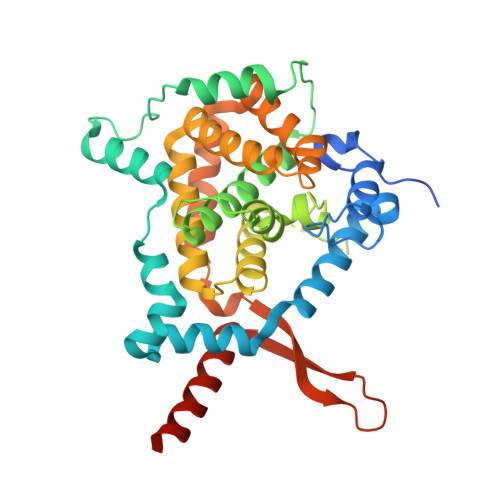
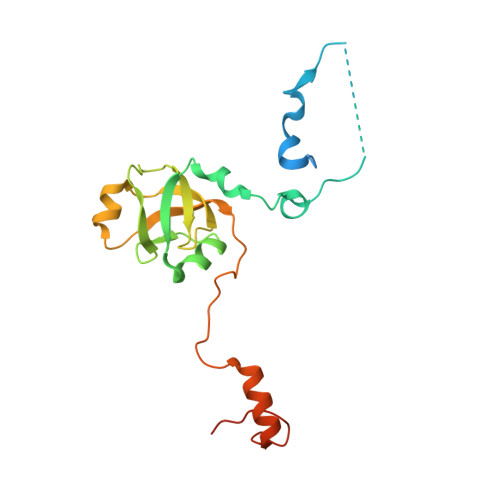
The H/Aca Rnp Assembly Factor Shq1 Functions as an RNA Mimic.
Walbott, H., Machado-Pinilla, R., Liger, D., Blaud, M., Rety, S., Grozdanov, P.N., Godin, K., Van Tilbeurgh, H., Varani, G., Meier, U.T., Leulliot, N.(2011) Genes Dev 25: 2398
- PubMed: 22085966
- DOI: https://doi.org/10.1101/gad.176834.111
- Primary Citation of Related Structures:
3ZUZ, 3ZV0 - PubMed Abstract:
SHQ1 is an essential assembly factor for H/ACA ribonucleoproteins (RNPs) required for ribosome biogenesis, pre-mRNA splicing, and telomere maintenance. SHQ1 binds dyskerin/NAP57, the catalytic subunit of human H/ACA RNPs, and this interaction is modulated by mutations causing X-linked dyskeratosis congenita. We report the crystal structure of the C-terminal domain of yeast SHQ1, Shq1p, and its complex with yeast dyskerin/NAP57, Cbf5p, lacking its catalytic domain. The C-terminal domain of Shq1p interacts with the RNA-binding domain of Cbf5p and, through structural mimicry, uses the RNA-protein-binding sites to achieve a specific protein-protein interface. We propose that Shq1p operates as a Cbf5p chaperone during RNP assembly by acting as an RNA placeholder, thereby preventing Cbf5p from nonspecific RNA binding before association with an H/ACA RNA and the other core RNP proteins.
Organizational Affiliation:
Institut de Biochimie et de Biophysique Moléculaire et Cellulaire, Université de Paris-Sud, Orsay Cedex, France.