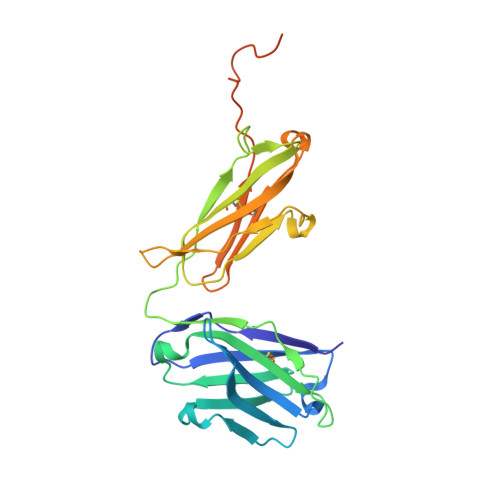
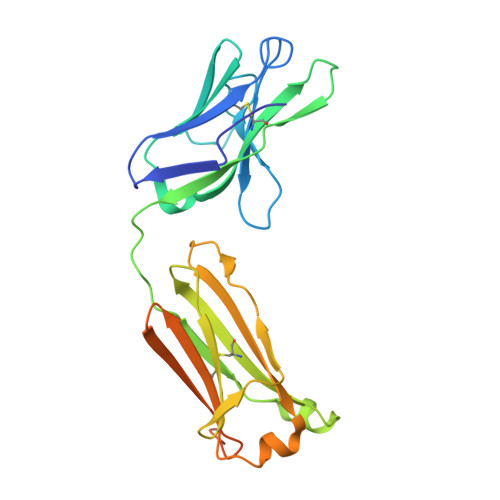
Role of Kappa>Lambda Light-Chain Constant-Domain Switch in the Structure and Functionality of A17 Reactibody
Ponomarenko, N.A., Chatziefthimiou, S.D., Kurkova, I.N., Mokrushina, Y.A., Stepanova, A.V., Smirnov, I.V., Avakyan, E.M., Bobik, T.V., Mamedov, A., Mitkevich, V.A., Belogurov, A.J., Fedorova, O.S., Dubina, M., Golovin, A., Lamzin, V.S., Friboulet, A., Makarov, A.A., Wilmanns, M., Gabibov, A.G.(2014) Acta Crystallogr D Biol Crystallogr 70: 708
- PubMed: 24598740
- DOI: https://doi.org/10.1107/S1399004713032446
- Primary Citation of Related Structures:
3ZL4 - PubMed Abstract:
The engineering of catalytic function in antibodies requires precise information on their structure. Here, results are presented that show how the antibody domain structure affects its functionality. The previously designed organophosphate-metabolizing reactibody A17 has been re-engineered by replacing its constant κ light chain by the λ chain (A17λ), and the X-ray structure of A17λ has been determined at 1.95 Å resolution. It was found that compared with A17κ the active centre of A17λ is displaced, stabilized and made more rigid owing to interdomain interactions involving the CDR loops from the VL and VH domains. These VL/VH domains also have lower mobility, as deduced from the atomic displacement parameters of the crystal structure. The antibody elbow angle is decreased to 126° compared with 138° in A17κ. These structural differences account for the subtle changes in catalytic efficiency and thermodynamic parameters determined with two organophosphate ligands, as well as in the affinity for peptide substrates selected from a combinatorial cyclic peptide library, between the A17κ and A17λ variants. The data presented will be of interest and relevance to researchers dealing with the design of antibodies with tailor-made functions.
Organizational Affiliation:
Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, ul. Miklukho-Maklaya 16/10, Moscow 117871, Russian Federation.