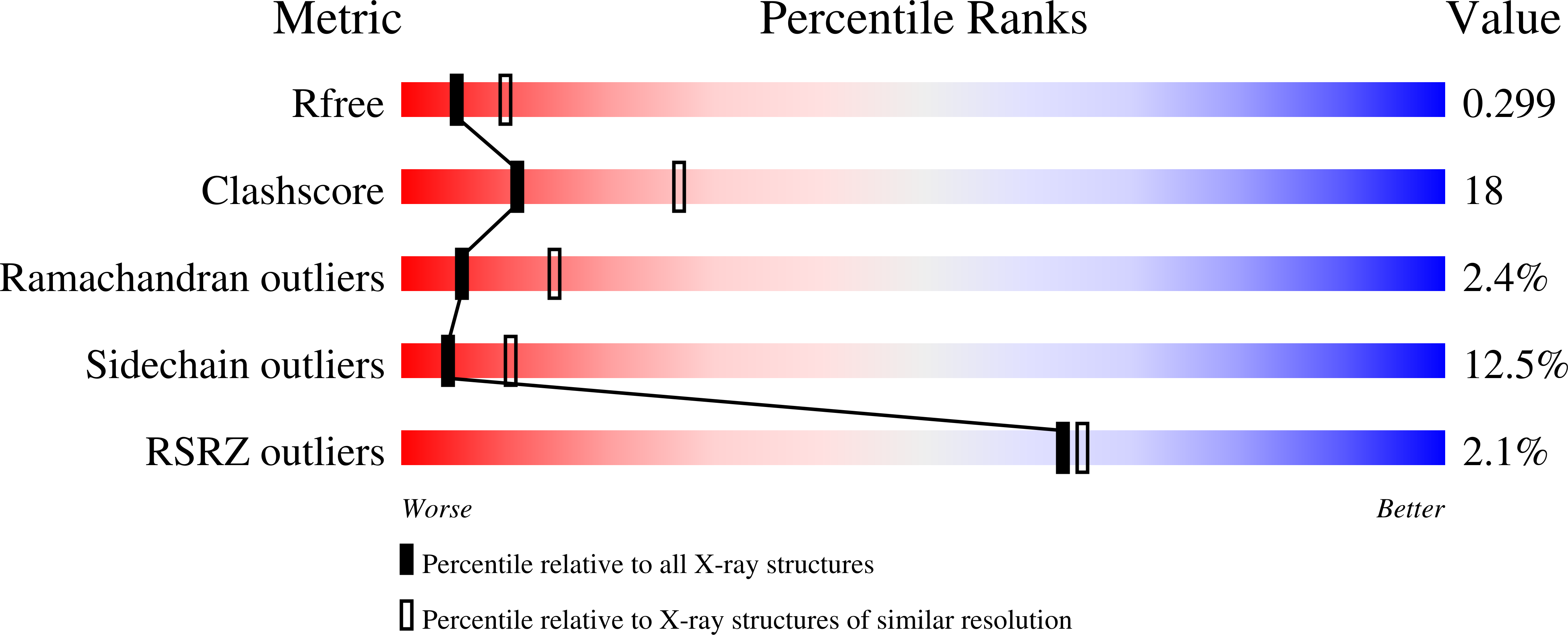
Molecular basis for the reverse reaction of African human trypanosomes glycerol kinase.
Balogun, E.O., Inaoka, D.K., Shiba, T., Kido, Y., Tsuge, C., Nara, T., Aoki, T., Honma, T., Tanaka, A., Inoue, M., Matsuoka, S., Michels, P.A., Kita, K., Harada, S.(2014) Mol Microbiol 94: 1315-1329
- PubMed: 25315291
- DOI: https://doi.org/10.1111/mmi.12831
- Primary Citation of Related Structures:
3WXI, 3WXJ, 3WXK, 3WXL - PubMed Abstract:
The glycerol kinase (GK) of African human trypanosomes is compartmentalized in their glycosomes. Unlike the host GK, which under physiological conditions catalyzes only the forward reaction (ATP-dependent glycerol phosphorylation), trypanosome GK can additionally catalyze the reverse reaction. In fact, owing to this unique reverse catalysis, GK is potentially essential for the parasites survival in the human host, hence a promising drug target. The mechanism of its reverse catalysis was unknown; therefore, it was not clear if this ability was purely due to its localization in the organelles or whether structure-based catalytic differences also contribute. To investigate this lack of information, the X-ray crystal structure of this protein was determined up to 1.90 Å resolution, in its unligated form and in complex with three natural ligands. These data, in conjunction with results from structure-guided mutagenesis suggests that the trypanosome GK is possibly a transiently autophosphorylating threonine kinase, with the catalytic site formed by non-conserved residues. Our results provide a series of structural peculiarities of this enzyme, and gives unexpected insight into the reverse catalysis mechanism. Together, they provide an encouraging molecular framework for the development of trypanosome GK-specific inhibitors, which may lead to the design of new and safer trypanocidal drug(s).
Organizational Affiliation:
Department of Applied Biology, Graduate School of Science and Technology, Kyoto Institute of Technology, Sakyo-ku, Kyoto, 606-8585, Japan; Department of Biomedical Chemistry, Graduate School of Medicine, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo, 113-0033, Japan; Department of Biochemistry, Ahmadu Bello University, Zaria, 2222, Nigeria.