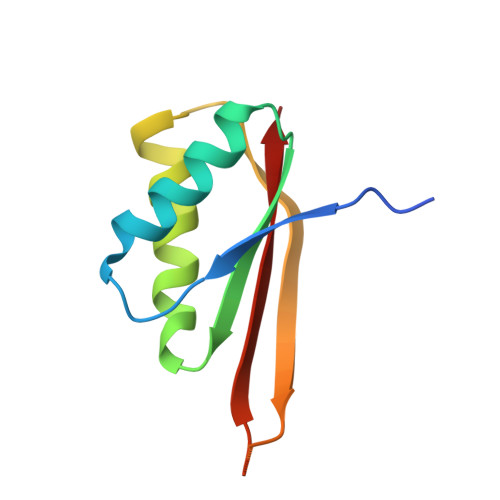
Biochemical and structural insights into RNA binding by Ssh10b, a member of the highly conserved Sac10b protein family in Archaea.
Guo, L., Ding, J., Guo, R., Hou, Y., Wang, D.C., Huang, L.(2014) J Biol Chem 289: 1478-1490
- PubMed: 24307170
- DOI: https://doi.org/10.1074/jbc.M113.521351
- Primary Citation of Related Structures:
3WBM - PubMed Abstract:
Proteins of the Sac10b family are highly conserved in Archaea. Ssh10b, a member of the Sac10b family from the hyperthermophilic crenarchaeon Sulfolobus shibatae, binds to RNA in vivo. Here we show that binding by Ssh10b destabilizes RNA secondary structure. Structural analysis of Ssh10b in complex with a 25-bp RNA duplex containing local distortions reveals that Ssh10b binds the two RNA strands symmetrically as a tetramer with each dimer bound asymmetrically to a single RNA strand. Amino acid residues involved in double-stranded RNA binding are similar, but non-identical, to those in dsDNA binding. The dimer-dimer interaction mediated by the intermolecular β-sheet appears to facilitate the destabilization of base pairing in the secondary structure of RNA. Our results suggest that proteins of the Sac10b family may play important roles in RNA transactions requiring destabilization of RNA secondary structure in Sulfolobus.
Organizational Affiliation:
From the State Key Laboratory of Microbial Resources, Institute of Microbiology and.