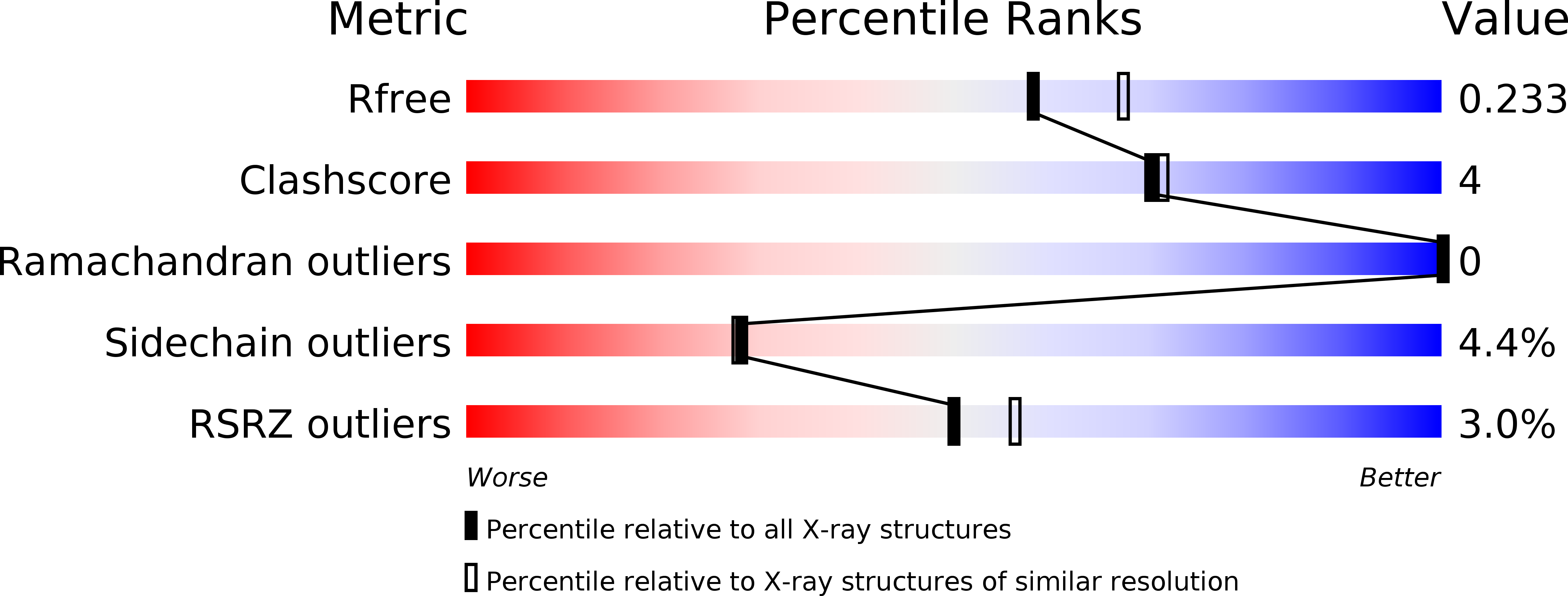
Insights into Duffy binding-like domains through the crystal structure and function of the merozoite surface protein MSPDBL2 from Plasmodium falciparum
Hodder, A.N., Czabotar, P.E., Uboldi, A.D., Clarke, O.B., Lin, C.S., Healer, J., Smith, B.J., Cowman, A.F.(2012) J Biol Chem 287: 32922-32939
- PubMed: 22843685
- DOI: https://doi.org/10.1074/jbc.M112.350504
- Primary Citation of Related Structures:
3VUU, 3VUV - PubMed Abstract:
Invasion of human red blood cells by Plasmodium falciparum involves interaction of the merozoite form through proteins on the surface coat. The erythrocyte binding-like protein family functions after initial merozoite interaction by binding via the Duffy binding-like (DBL) domain to receptors on the host red blood cell. The merozoite surface proteins DBL1 and -2 (PfMSPDBL1 and PfMSPDBL2) (PF10_0348 and PF10_0355) are extrinsically associated with the merozoite, and both have a DBL domain in each protein. We expressed and refolded recombinant DBL domains for PfMSPDBL1 and -2 and show they are functional. The red cell binding characteristics of these domains were shown to be similar to full-length forms of these proteins isolated from parasite cultures. Futhermore, metal cofactors were found to enhance the binding of both the DBL domains and the parasite-derived full-length proteins to erythrocytes, which has implications for receptor binding of other DBL-containing proteins in Plasmodium spp. We solved the structure of the erythrocyte-binding DBL domain of PfMSPDBL2 to 2.09 Å resolution and modeled that of PfMSPDBL1, revealing a canonical DBL fold consisting of a boomerang shaped α-helical core formed from three subdomains. PfMSPDBL2 is highly polymorphic, and mapping of these mutations shows they are on the surface, predominantly in the first two domains. For both PfMSPDBL proteins, polymorphic variation spares the cleft separating domains 1 and 2 from domain 3, and the groove between the two major helices of domain 3 extends beyond the cleft, indicating these regions are functionally important and are likely to be associated with the binding of a receptor on the red blood cell.
Organizational Affiliation:
The Walter and Eliza Hall Institute of Medical Research, Melbourne 3052, Australia. hodder@wehi.edu.au