Crystal structure of a influenza A virus (A/Aichi/2/1968 H3N2) hemagglutinin in C2 space group
Yasutake, Y., Suzuki, T., Kawaguchi, A., Nobusawa, E.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hemagglutinin HA1 chain | 329 | Influenza A virus (A/Aichi/2/1968(H3N2)) | Mutation(s): 0 | 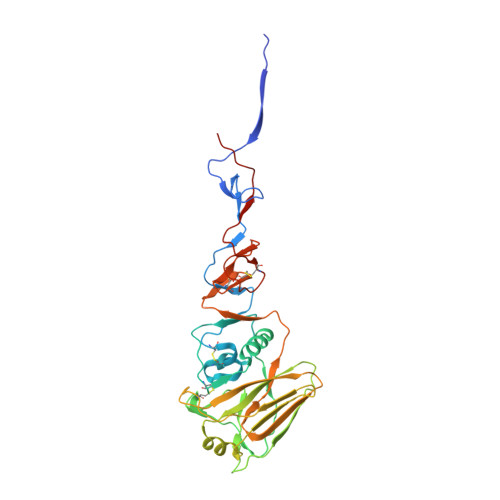 | |
UniProt | |||||
Find proteins for P03437 (Influenza A virus (strain A/Aichi/2/1968 H3N2)) Explore P03437 Go to UniProtKB: P03437 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P03437 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hemagglutinin HA2 chain | 175 | Influenza A virus (A/Aichi/2/1968(H3N2)) | Mutation(s): 0 | 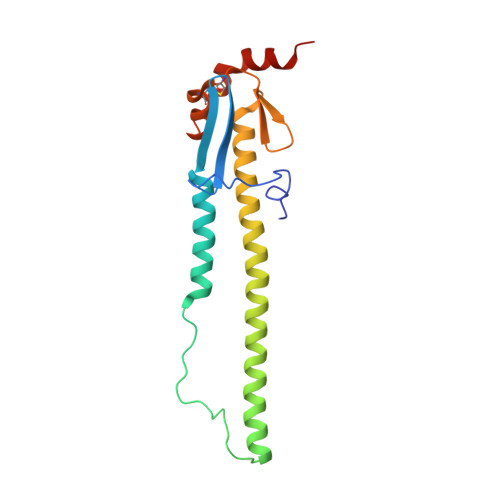 | |
UniProt | |||||
Find proteins for P03437 (Influenza A virus (strain A/Aichi/2/1968 H3N2)) Explore P03437 Go to UniProtKB: P03437 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P03437 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | G, J, M | 9 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G27919IH GlyCosmos: G27919IH GlyGen: G27919IH | |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | H, K, N | 2 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G42666HT GlyCosmos: G42666HT GlyGen: G42666HT | |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | I, L, O | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G22573RC GlyCosmos: G22573RC GlyGen: G22573RC | |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | P [auth A] Q [auth B] R [auth C] S [auth D] T [auth E] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 170.016 | α = 90 |
| b = 98.13 | β = 113.11 |
| c = 144.776 | γ = 90 |
| Software Name | Purpose |
|---|---|
| ADSC | data collection |
| MOLREP | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |