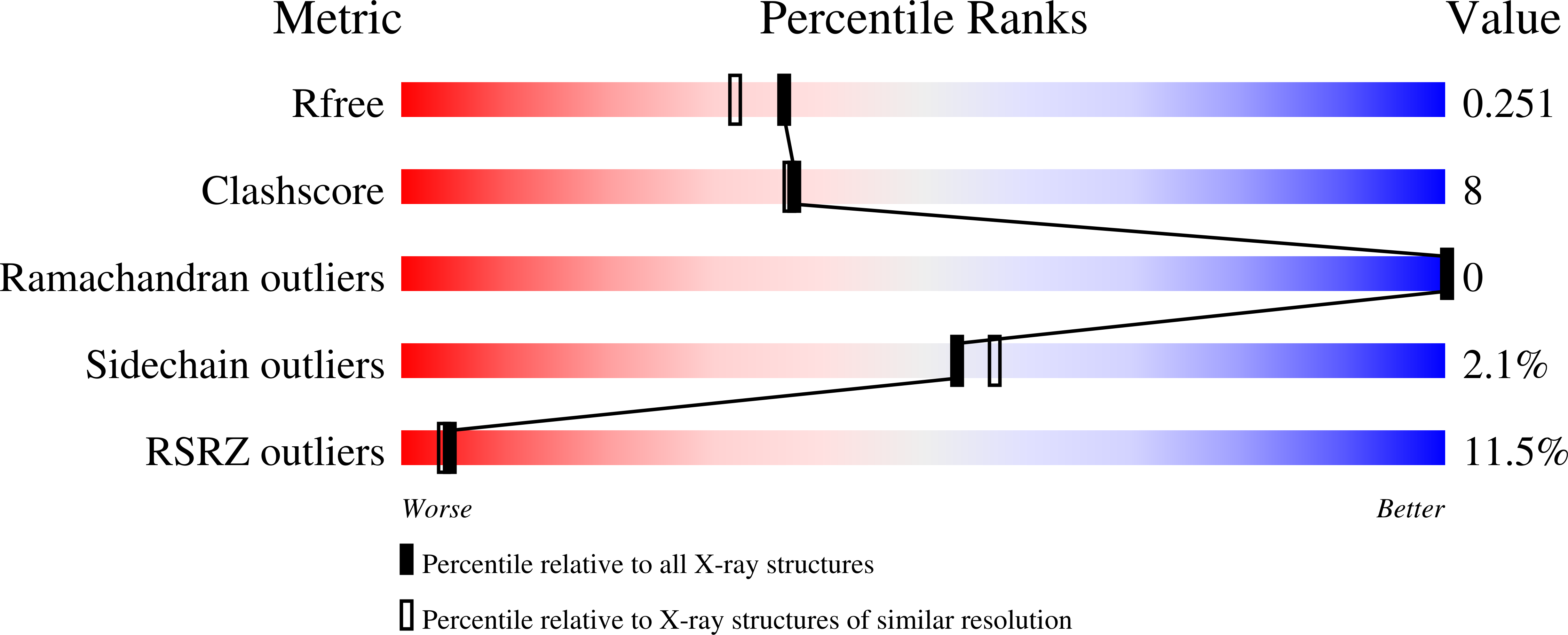
The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold
Zhang, A.P.P., Bornholdt, Z.A., Liu, T., Abelson, D.M., Lee, D.E., Li, S., Woods Jr., V.L., Saphire, E.O.(2012) PLoS Pathog 8: e1002550-e1002550
- PubMed: 22383882
- DOI: https://doi.org/10.1371/journal.ppat.1002550
- Primary Citation of Related Structures:
3VNE, 3VNF, 4D9O - PubMed Abstract:
Ebolaviruses cause hemorrhagic fever with up to 90% lethality and in fatal cases, are characterized by early suppression of the host innate immune system. One of the proteins likely responsible for this effect is VP24. VP24 is known to antagonize interferon signaling by binding host karyopherin α proteins, thereby preventing them from transporting the tyrosine-phosphorylated transcription factor STAT1 to the nucleus. Here, we report that VP24 binds STAT1 directly, suggesting that VP24 can suppress at least two distinct branches of the interferon pathway. Here, we also report the first crystal structures of VP24, derived from different species of ebolavirus that are pathogenic (Sudan) and nonpathogenic to humans (Reston). These structures reveal that VP24 has a novel, pyramidal fold. A site on a particular face of the pyramid exhibits reduced solvent exchange when in complex with STAT1. This site is above two highly conserved pockets in VP24 that contain key residues previously implicated in virulence. These crystal structures and accompanying biochemical analysis map differences between pathogenic and nonpathogenic viruses, offer templates for drug design, and provide the three-dimensional framework necessary for biological dissection of the many functions of VP24 in the virus life cycle.
Organizational Affiliation:
Department of Immunology and Microbial Science, The Scripps Research Institute, La Jolla, California, USA.