Crystal and solution structures of methyltransferase RsmH provide basis for methylation of C1402 in 16S rRNA.
Wei, Y., Zhang, H., Gao, Z.Q., Wang, W.J., Shtykova, E.V., Xu, J.H., Liu, Q.S., Dong, Y.H.(2012) J Struct Biol 179: 29-40
- PubMed: 22561317
- DOI: https://doi.org/10.1016/j.jsb.2012.04.011
- Primary Citation of Related Structures:
3TKA - PubMed Abstract:
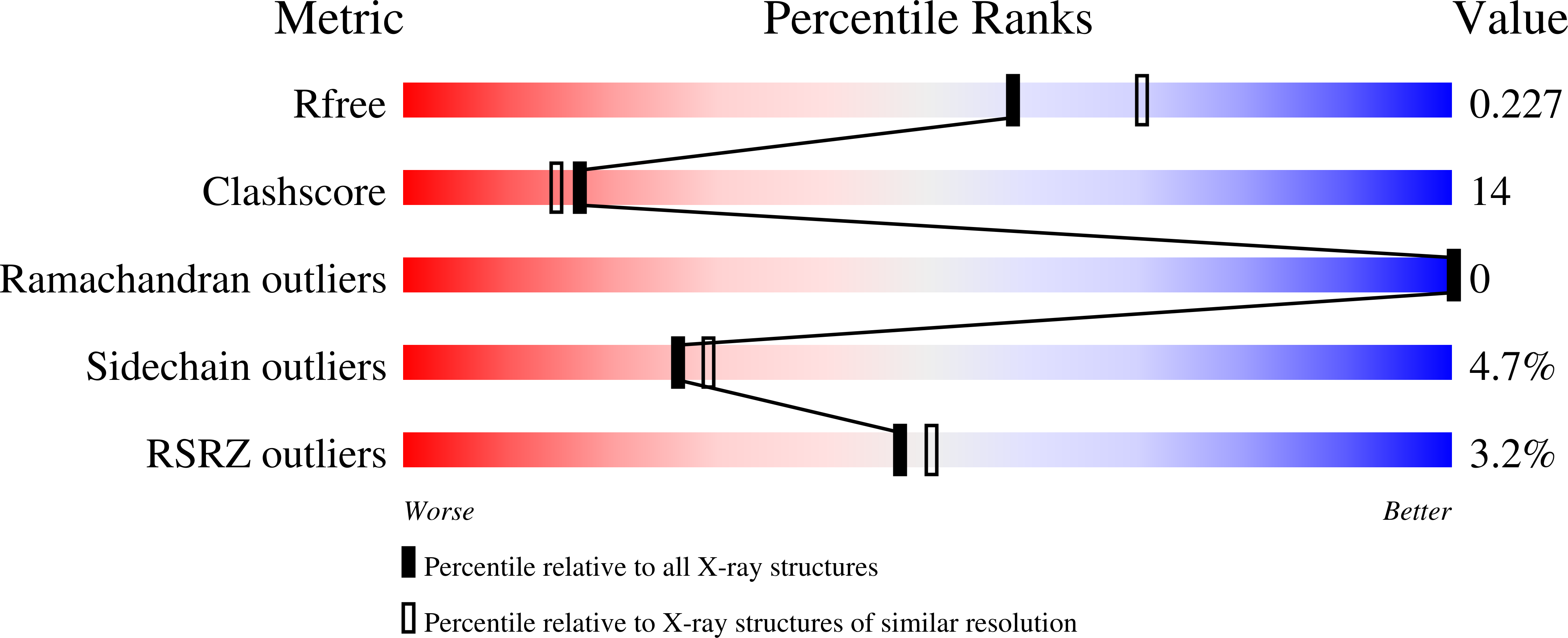
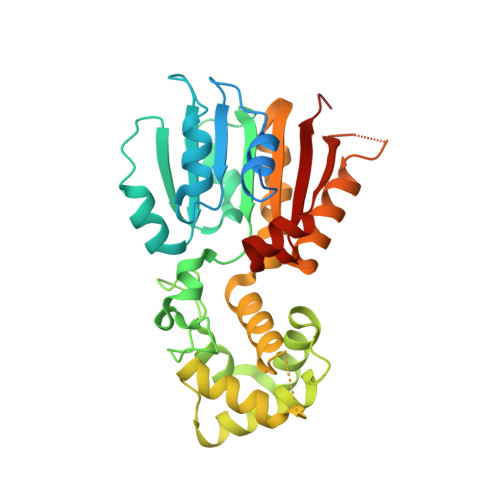
RsmH is a specific AdoMet-dependent methyltransferase (MTase) responsible for N(4)-methylation of C1402 in 16S rRNA and conserved in almost all species of bacteria. The methylcytidine interacts with the P-site codon of the mRNA and increases ribosomal decoding fidelity. In this study, high resolution crystal structure (2.25Å) of Escherichia coli RsmH in complex with AdoMet and cytidine (the putative rRNA binding site) was determined. The structural analysis demonstrated that the complex consists of two distinct but structurally related domains: the typical MTase domain and the putative substrate recognition and binding domain. A deep pocket was found in the conserved AdoMet binding domain. It was also found that the cytidine bound far from AdoMet with the distance of 25.9Å. It indicates that the complex is not in a catalytically active state, and structural rearrangement of RsmH or the nucleotides neighboring C1402 may be necessary to trigger catalysis. Although there is only one molecule in the asymmetric unit of the crystals, RsmH can form a compact dimer across a crystallographic twofold axis. Further analysis of RsmH by small-angle X-ray scattering (SAXS) also revealed the dimer in solution, but with a more flexible conformation than that in crystal, likely resulting from the absence of the substrate. It implies that an active status of RsmH in vivo is achieved by a formation of the dimeric architecture. In general, crystal and solution structural analysis provides new information on the mechanism of the methylation of the fine-tuning ribosomal decoding center by the RsmH.
Organizational Affiliation:
School of Life Sciences, University of Science and Technology of China, Hefei 230027, People's Republic of China.