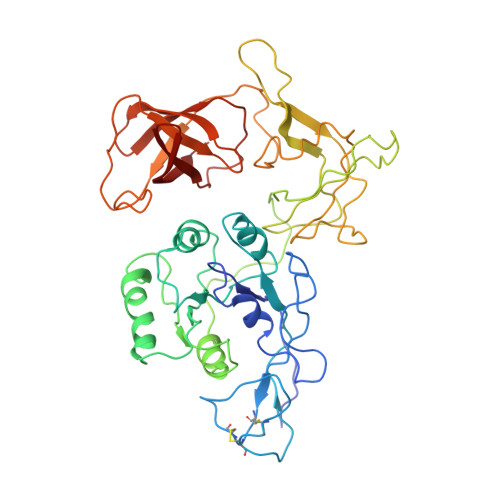
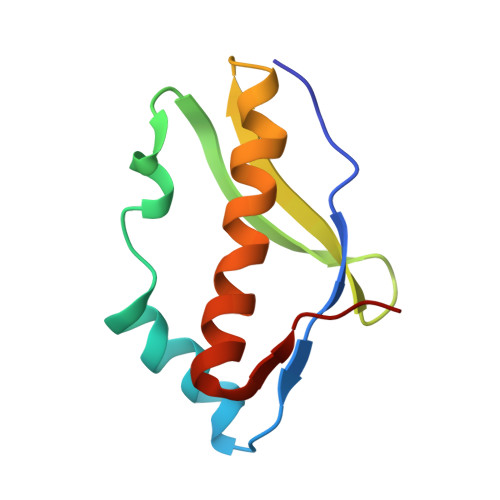
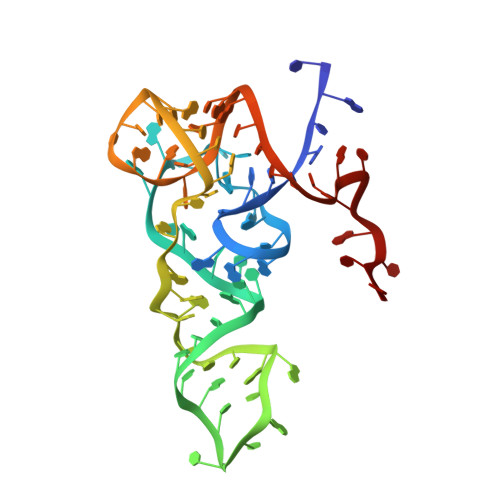
Crystal structure of the archaeal translation initiation factor 2 in complex with a GTP analogue and Met-tRNAf(Met.)
Stolboushkina, E., Nikonov, S., Zelinskaya, N., Arkhipova, V., Nikulin, A., Garber, M., Nikonov, O.(2013) J Mol Biol 425: 989-998
- PubMed: 23291527
- DOI: https://doi.org/10.1016/j.jmb.2012.12.023
- Primary Citation of Related Structures:
3QSY - PubMed Abstract:
Heterotrimeric aIF2αβγ (archaeal homologue of the eukaryotic translation initiation factor 2) in its GTP-bound form delivers Met-tRNAi(Met) to the small ribosomal subunit. It is known that the heterodimer containing the GTP-bound γ subunit and domain 3 of the α subunit of aIF2 is required for the formation of a stable complex with Met-tRNAi. Here, the crystal structure of an incomplete ternary complex including aIF2αD3γ⋅GDPNP⋅Met-tRNAf(Met) has been solved at 3.2Å resolution. This structure is in good agreement with biochemical and hydroxyl radical probing data. The analysis of the complex shows that despite the structural similarity of aIF2γ and the bacterial translation elongation factor EF-Tu, their modes of tRNA binding are very different. Remarkably, the recently published 5.0-Å-resolution structure of almost the same ternary initiation complex differs dramatically from the structure presented. Reasons for this discrepancy are discussed.
Organizational Affiliation:
Institute of Protein Research, Russian Academy of Sciences, 142290, Pushchino, Moscow Region, Russian Federation.