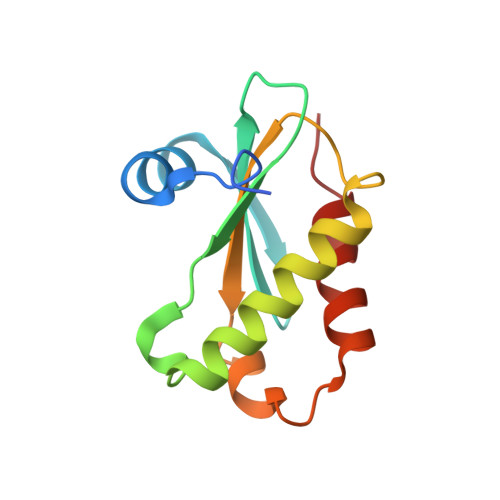
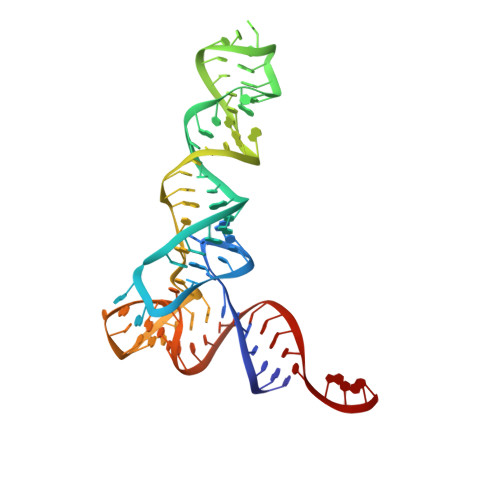
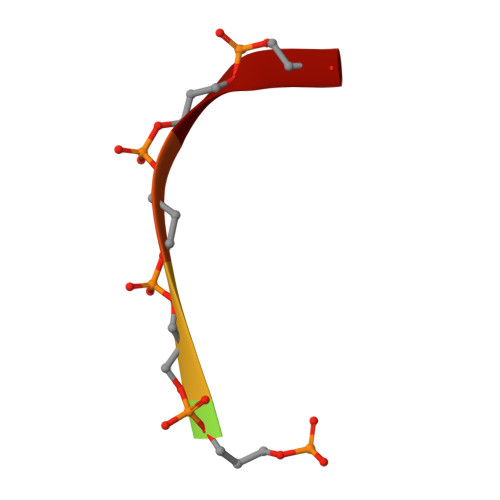
Structure of a Bacterial Ribonuclease P Holoenzyme in Complex with tRNA.
Reiter, N.J., Osterman, A., Torres-Larios, A., Swinger, K.K., Pan, T., Mondragon, A.(2010) Nature 468: 784-789
- PubMed: 21076397
- DOI: https://doi.org/10.1038/nature09516
- Primary Citation of Related Structures:
3Q1Q, 3Q1R - PubMed Abstract:
Ribonuclease (RNase) P is the universal ribozyme responsible for 5'-end tRNA processing. We report the crystal structure of the Thermotoga maritima RNase P holoenzyme in complex with tRNA(Phe). The 154 kDa complex consists of a large catalytic RNA (P RNA), a small protein cofactor and a mature tRNA. The structure shows that RNA-RNA recognition occurs through shape complementarity, specific intermolecular contacts and base-pairing interactions. Soaks with a pre-tRNA 5' leader sequence with and without metal help to identify the 5' substrate path and potential catalytic metal ions. The protein binds on top of a universally conserved structural module in P RNA and interacts with the leader, but not with the mature tRNA. The active site is composed of phosphate backbone moieties, a universally conserved uridine nucleobase, and at least two catalytically important metal ions. The active site structure and conserved RNase P-tRNA contacts suggest a universal mechanism of catalysis by RNase P.
Organizational Affiliation:
Department of Molecular Biosciences, Northwestern University, Evanston, Illinois 60208, USA.