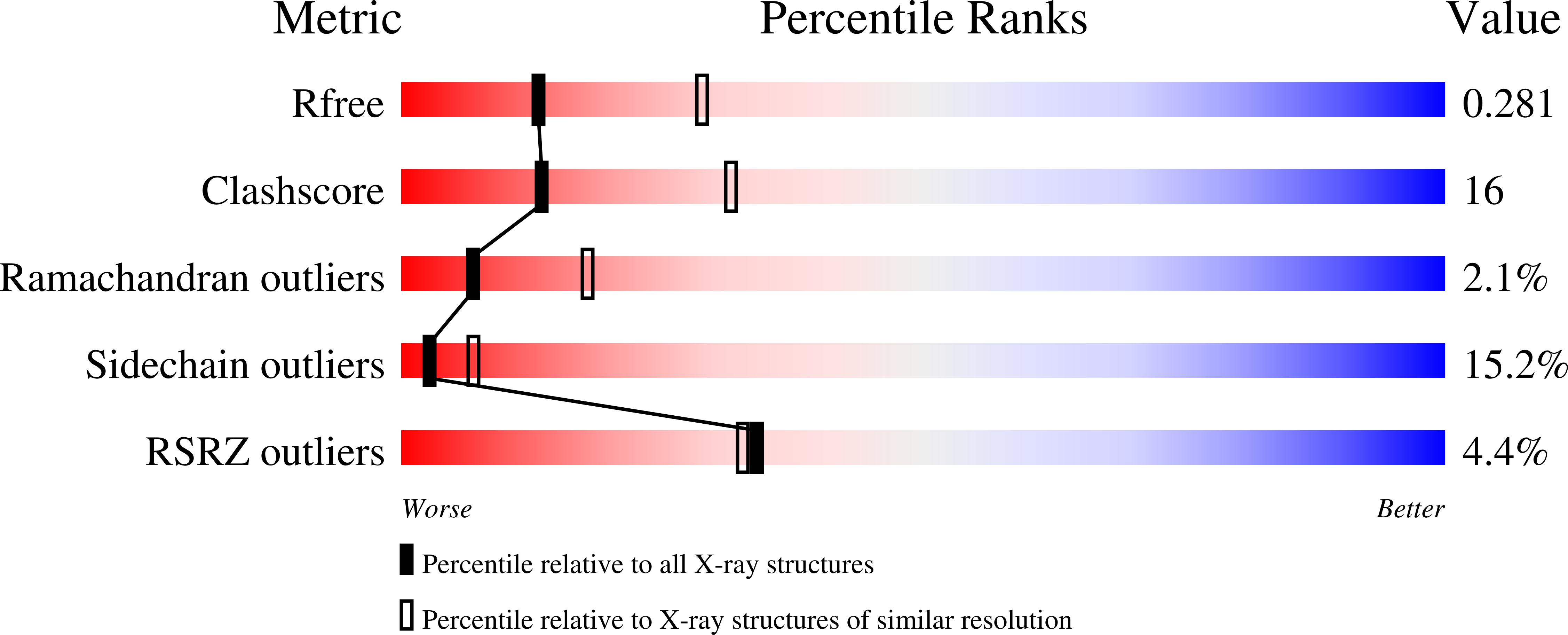
Crystal Structure of Bacillus anthracis Phosphoglucosamine Mutase, an Enzyme in the Peptidoglycan Biosynthetic Pathway.
Mehra-Chaudhary, R., Mick, J., Beamer, L.J.(2011) J Bacteriol 193: 4081-4087
- PubMed: 21685296
- DOI: https://doi.org/10.1128/JB.00418-11
- Primary Citation of Related Structures:
3PDK - PubMed Abstract:
Phosphoglucosamine mutase (PNGM) is an evolutionarily conserved bacterial enzyme that participates in the cytoplasmic steps of peptidoglycan biosynthesis. As peptidoglycan is essential for bacterial survival and is absent in humans, enzymes in this pathway have been the focus of intensive inhibitor design efforts. Many aspects of the structural biology of the peptidoglycan pathway have been elucidated, with the exception of the PNGM structure. We present here the crystal structure of PNGM from the human pathogen and bioterrorism agent Bacillus anthracis. The structure reveals key residues in the large active site cleft of the enzyme which likely have roles in catalysis and specificity. A large conformational change of the C-terminal domain of PNGM is observed when comparing two independent molecules in the crystal, shedding light on both the apo- and ligand-bound conformers of the enzyme. Crystal packing analyses and dynamic light scattering studies suggest that the enzyme is a dimer in solution. Multiple sequence alignments show that residues in the dimer interface are conserved, suggesting that many PNGM enzymes adopt this oligomeric state. This work lays the foundation for the development of inhibitors for PNGM enzymes from human pathogens.
Organizational Affiliation:
Biochemistry Department, University of Missouri, Columbia, MO 65211, USA.