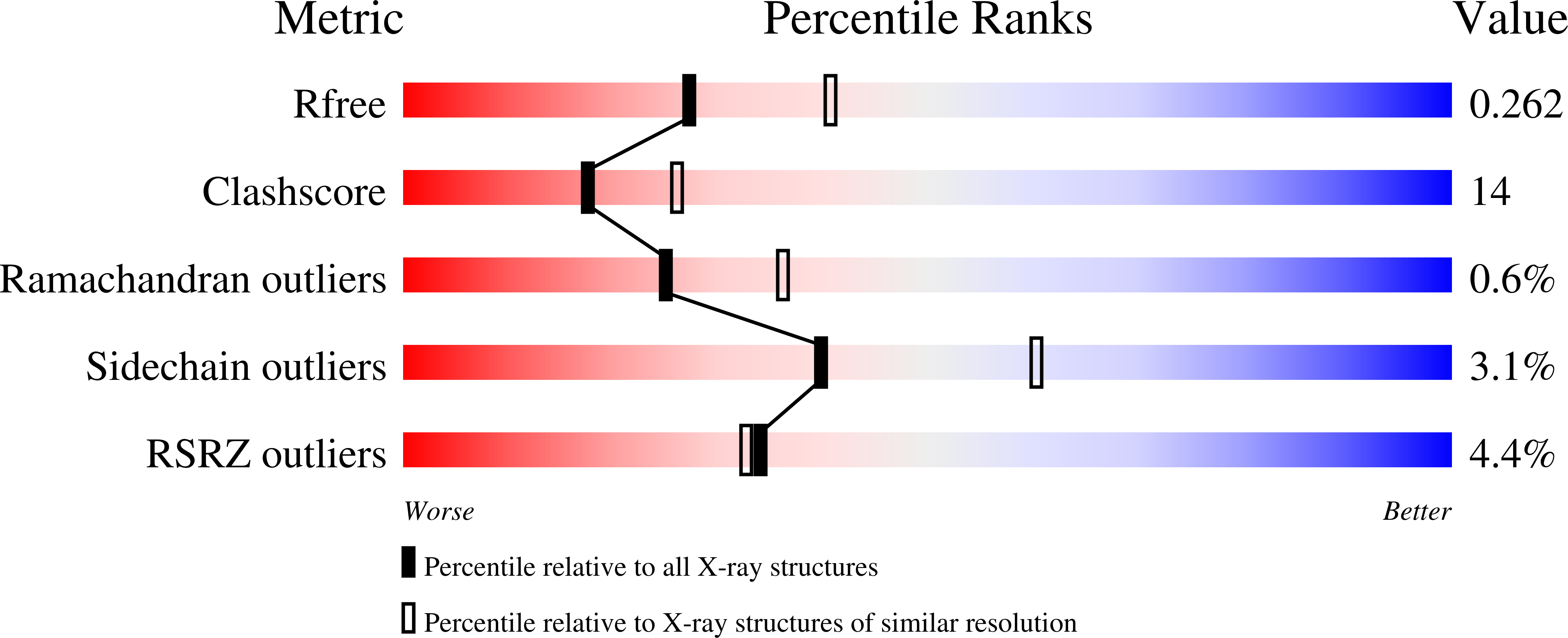
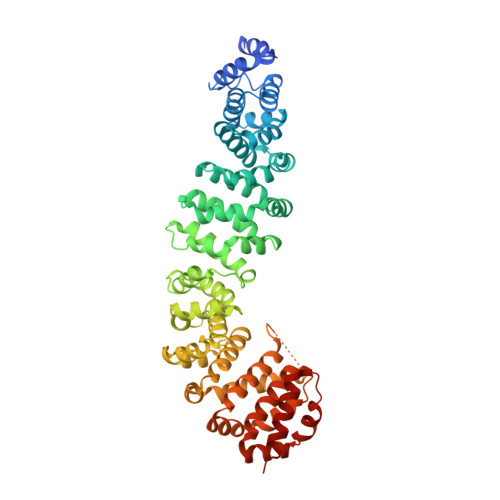
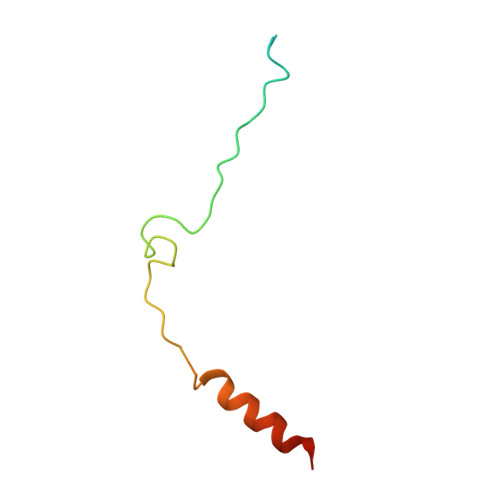
Biochemical and structural characterization of beta-catenin interactions with nonphosphorylated and CK2-phosphorylated Lef-1.
Sun, J., Weis, W.I.(2011) J Mol Biol 405: 519-530
- PubMed: 21075118
- DOI: https://doi.org/10.1016/j.jmb.2010.11.010
- Primary Citation of Related Structures:
3OUW, 3OUX - PubMed Abstract:
In the Wnt/β-catenin signaling pathway, β-catenin activates target genes through its interactions with the T-cell factor/lymphoid enhancer-binding factor (TCF/Lef) family of transcription factors. The crystal structures of complexes between the β-catenin armadillo domain and the Lef-1 N-terminal domain show that the overall conformation and many of the interactions are similar to other published structures of TCFs bound to β-catenin. However, a second salt bridge in other TCF-β-catenin structures is absent in the structure of β-catenin-Lef-1 complex, indicating that this feature is not obligatory for β-catenin binding. Casein kinase II (CK2) has been shown to act as a positive regulator of Wnt signaling, and Lef-1 is a substrate of CK2. In vitro phosphorylation of purified Lef-1 was used to examine the effect of CK2 on the interaction of Lef-1 with β-catenin. Mass spectrometry data show that CK2 phosphorylation of Lef-1 N-terminal domain results in a single phosphorylation site at Ser40. Isothermal titration calorimetry revealed that β-catenin binds to nonphosphorylated or CK2-phosphorylated Lef-1 with the same affinity, which is consistent with the absence of phospho-Ser40 interactions in the crystal structure of phosphorylated Lef-1 N-terminal domain bound to β-catenin. These data indicate that the effect of CK2 on the Wnt/β-catenin pathway does not appear to be at the level of the Lef-1-β-catenin interaction.
Organizational Affiliation:
Department of Structural Biology, Stanford University School of Medicine,Stanford, CA 94305-5126, USA.