Trypanosoma cruzi trans-sialidase in complex with a neutralizing antibody: structure/function studies towards the rational design of inhibitors.
Buschiazzo, A., Muia, R., Larrieux, N., Pitcovsky, T., Mucci, J., Campetella, O.(2012) PLoS Pathog 8: e1002474-e1002474
- PubMed: 22241998
- DOI: https://doi.org/10.1371/journal.ppat.1002474
- Primary Citation of Related Structures:
3OPZ - PubMed Abstract:
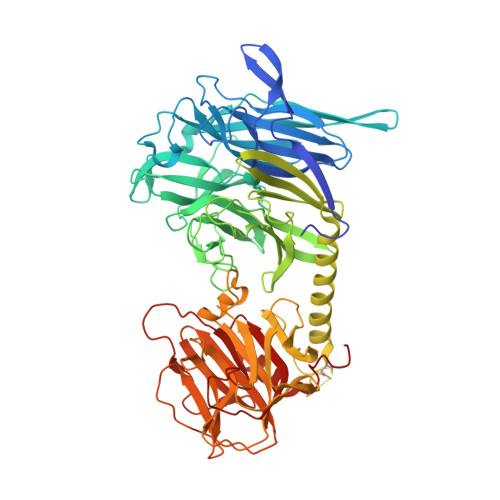
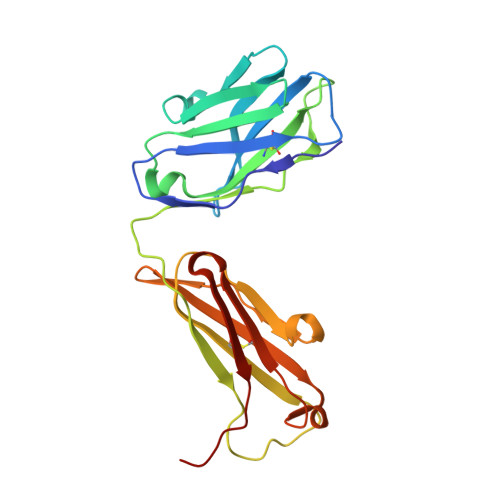
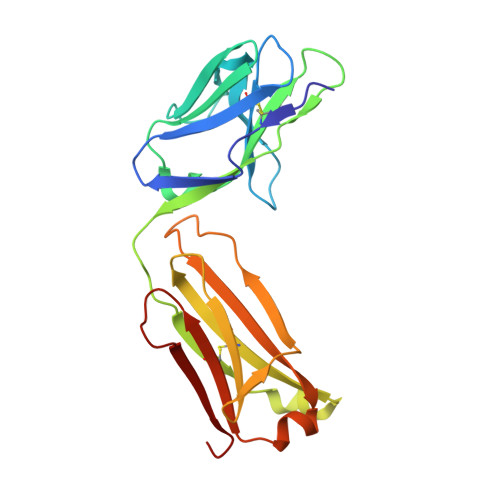
Trans-sialidase (TS), a virulence factor from Trypanosoma cruzi, is an enzyme playing key roles in the biology of this protozoan parasite. Absent from the mammalian host, it constitutes a potential target for the development of novel chemotherapeutic drugs, an urgent need to combat Chagas' disease. TS is involved in host cell invasion and parasite survival in the bloodstream. However, TS is also actively shed by the parasite to the bloodstream, inducing systemic effects readily detected during the acute phase of the disease, in particular, hematological alterations and triggering of immune cells apoptosis, until specific neutralizing antibodies are elicited. These antibodies constitute the only known submicromolar inhibitor of TS's catalytic activity. We now report the identification and detailed characterization of a neutralizing mouse monoclonal antibody (mAb 13G9), recognizing T. cruzi TS with high specificity and subnanomolar affinity. This mAb displays undetectable association with the T. cruzi superfamily of TS-like proteins or yet with the TS-related enzymes from Trypanosoma brucei or Trypanosoma rangeli. In immunofluorescence assays, mAb 13G9 labeled 100% of the parasites from the infective trypomastigote stage. This mAb also reduces parasite invasion of cultured cells and strongly inhibits parasite surface sialylation. The crystal structure of the mAb 13G9 antigen-binding fragment in complex with the globular region of T. cruzi TS was determined, revealing detailed molecular insights of the inhibition mechanism. Not occluding the enzyme's catalytic site, the antibody performs a subtle action by inhibiting the movement of an assisting tyrosine (Y₁₁₉), whose mobility is known to play a key role in the trans-glycosidase mechanism. As an example of enzymatic inhibition involving non-catalytic residues that occupy sites distal from the substrate-binding pocket, this first near atomic characterization of a high affinity inhibitory molecule for TS provides a rational framework for novel strategies in the design of chemotherapeutic compounds.
Organizational Affiliation:
Institut Pasteur de Montevideo, Unit of Protein Crystallography, Montevideo, Uruguay. alebus@pasteur.edu.uy