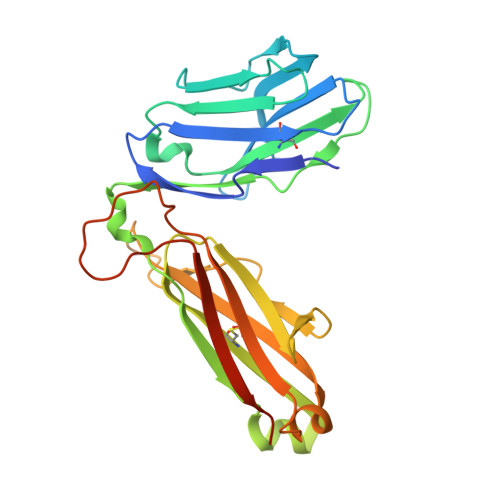
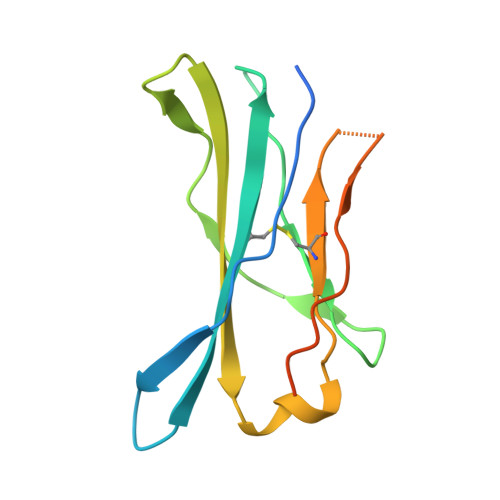
The structural basis for autonomous dimerization of the pre-T-cell antigen receptor
Pang, S.S., Berry, R., Chen, Z., Kjer-Nielsen, L., Perugini, M.A., King, G.F., Wang, C., Chew, S.H., La Gruta, N.L., Williams, N.K., Beddoe, T., Tiganis, T., Cowieson, N.P., Godfrey, D.I., Purcell, A.W., Wilce, M.C.J., McCluskey, J., Rossjohn, J.(2010) Nature 467: 844-848
- PubMed: 20944746
- DOI: https://doi.org/10.1038/nature09448
- Primary Citation of Related Structures:
3OF6 - PubMed Abstract:
The pre-T-cell antigen receptor (pre-TCR), expressed by immature thymocytes, has a pivotal role in early T-cell development, including TCR β-selection, survival and proliferation of CD4(-)CD8(-) double-negative thymocytes, and subsequent αβ T-cell lineage differentiation. Whereas αβTCR ligation by the peptide-loaded major histocompatibility complex initiates T-cell signalling, pre-TCR-induced signalling occurs by means of a ligand-independent dimerization event. The pre-TCR comprises an invariant α-chain (pre-Tα) that pairs with any TCR β-chain (TCRβ) following successful TCR β-gene rearrangement. Here we provide the basis of pre-Tα-TCRβ assembly and pre-TCR dimerization. The pre-Tα chain comprised a single immunoglobulin-like domain that is structurally distinct from the constant (C) domain of the TCR α-chain; nevertheless, the mode of association between pre-Tα and TCRβ mirrored that mediated by the Cα-Cβ domains of the αβTCR. The pre-TCR had a propensity to dimerize in solution, and the molecular envelope of the pre-TCR dimer correlated well with the observed head-to-tail pre-TCR dimer. This mode of pre-TCR dimerization enabled the pre-Tα domain to interact with the variable (V) β domain through residues that are highly conserved across the Vβ and joining (J) β gene families, thus mimicking the interactions at the core of the αβTCR's Vα-Vβ interface. Disruption of this pre-Tα-Vβ dimer interface abrogated pre-TCR dimerization in solution and impaired pre-TCR expression on the cell surface. Accordingly, we provide a mechanism of pre-TCR self-association that allows the pre-Tα chain to simultaneously 'sample' the correct folding of both the V and C domains of any TCR β-chain, regardless of its ultimate specificity, which represents a critical checkpoint in T-cell development. This unusual dual-chaperone-like sensing function of pre-Tα represents a unique mechanism in nature whereby developmental quality control regulates the expression and signalling of an integral membrane receptor complex.
Organizational Affiliation:
The Protein Crystallography Unit, Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria 3800, Australia.