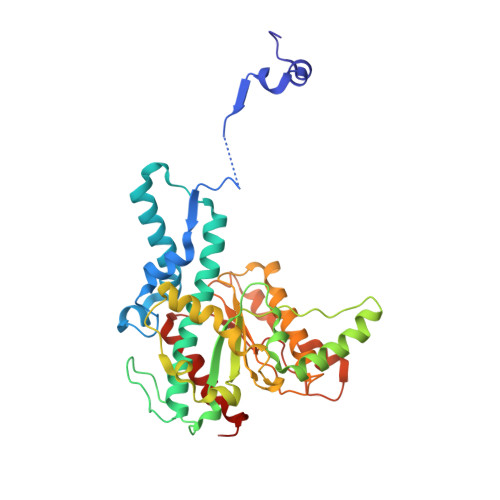
Domain swapping in allosteric modulation of DNA specificity.
Park, C.K., Joshi, H.K., Agrawal, A., Ghare, M.I., Little, E.J., Dunten, P.W., Bitinaite, J., Horton, N.C.(2010) PLoS Biol 8: e1000554-e1000554
- PubMed: 21151881
- DOI: https://doi.org/10.1371/journal.pbio.1000554
- Primary Citation of Related Structures:
3MQ6 - PubMed Abstract:
SgrAI is a type IIF restriction endonuclease that cuts an unusually long recognition sequence and exhibits allosteric self-modulation of cleavage activity and sequence specificity. Previous studies have shown that DNA bound dimers of SgrAI oligomerize into an activated form with higher DNA cleavage rates, although previously determined crystal structures of SgrAI bound to DNA show only the DNA bound dimer. A new crystal structure of the type II restriction endonuclease SgrAI bound to DNA and Ca(2+) is now presented, which shows the close association of two DNA bound SgrAI dimers. This tetrameric form is unlike those of the homologous enzymes Cfr10I and NgoMIV and is formed by the swapping of the amino-terminal 24 amino acid residues. Two mutations predicted to destabilize the swapped form of SgrAI, P27W and P27G, have been made and shown to eliminate both the oligomerization of the DNA bound SgrAI dimers as well as the allosteric stimulation of DNA cleavage by SgrAI. A mechanism involving domain swapping is proposed to explain the unusual allosteric properties of SgrAI via association of the domain swapped tetramer of SgrAI bound to DNA into higher order oligomers.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Arizona, Tucson, Arizona, United States of America.