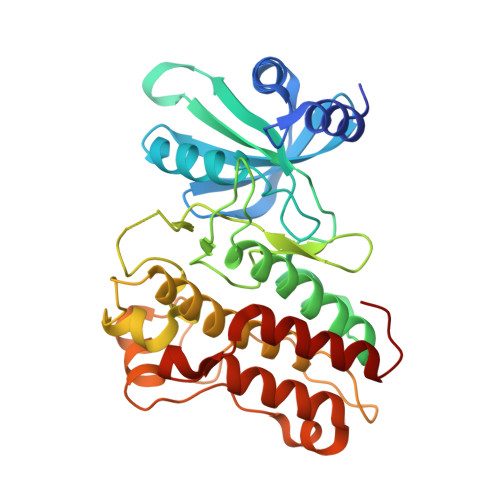
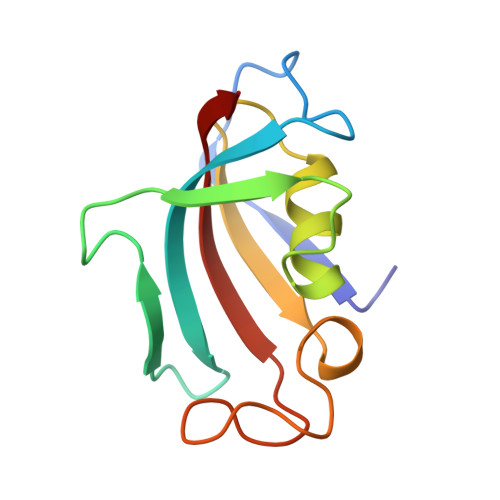
Crystal structure of the cytoplasmic domain of the bone morphogenetic protein receptor type-1B (BMPR1B) in complex with FKBP12 and LDN-193189
Chaikuad, A., Sanvitale, C., Mahajan, P., Daga, N., Cooper, C., Krojer, T., Alfano, I., Knapp, S., von Delft, F., Weigelt, J., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Bullock, A., Structural Genomics Consortium (SGC)To be published.