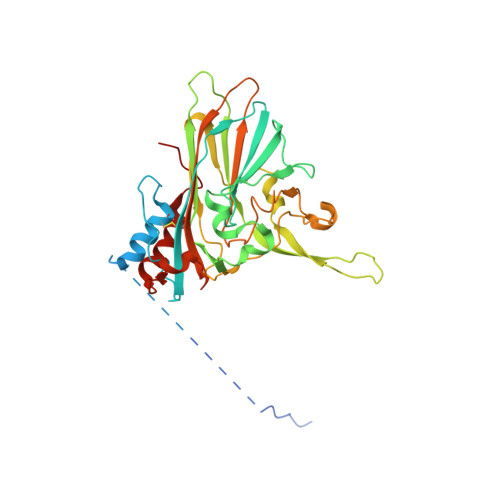
Structure and function of a genetically engineered mimic of a nonenveloped virus entry intermediate.
Banerjee, M., Speir, J.A., Kwan, M.H., Huang, R., Aryanpur, P.P., Bothner, B., Johnson, J.E.(2010) J Virol 84: 4737-4746
- PubMed: 20164221
- DOI: https://doi.org/10.1128/JVI.02670-09
- Primary Citation of Related Structures:
3LOB - PubMed Abstract:
Divalent metal ions are components of numerous icosahedral virus capsids. Flock House virus (FHV), a small RNA virus of the family Nodaviridae, was utilized as an accessible model system with which to address the effects of metal ions on capsid structure and on the biology of virus-host interactions. Mutations at the calcium-binding sites affected FHV capsid stability and drastically reduced virus infectivity, without altering the overall architecture of the capsid. The mutations also altered the conformation of gamma, a membrane-disrupting, virus-encoded peptide usually sequestered inside the capsid, by increasing its exposure under neutral pH conditions. Our data demonstrate that calcium binding is essential for maintaining a pH-based control on gamma exposure and host membrane disruption, and they reveal a novel rationale for the metal ion requirement during virus entry and infectivity. In the light of the phenotypes displayed by a calcium site mutant of FHV, we suggest that this mutant corresponds to an early entry intermediate formed in the endosomal pathway.
Organizational Affiliation:
Department of Molecular Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.