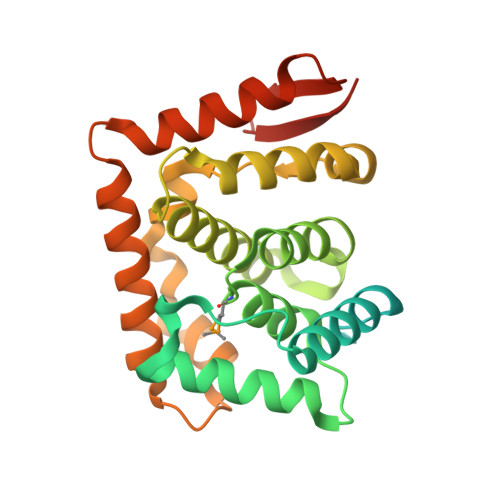
Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Sampathkumar, P., Gheyi, T., Miller, S.A., Bain, K.T., Dickey, M., Bonanno, J.B., Kim, S.J., Phillips, J., Pieper, U., Fernandez-Martinez, J., Franke, J.D., Martel, A., Tsuruta, H., Atwell, S., Thompson, D.A., Emtage, J.S., Wasserman, S.R., Rout, M.P., Sali, A., Sauder, J.M., Burley, S.K.(2011) Proteins 79: 1672-1677
- PubMed: 21365675
- DOI: https://doi.org/10.1002/prot.22973
- Primary Citation of Related Structures:
3KFO - PubMed Abstract:
Nuclear pore complexes (NPCs), responsible for the nucleo-cytoplasmic exchange of proteins and nucleic acids, are dynamic macromolecular assemblies forming an eight-fold symmetric co-axial ring structure. Yeast ( Saccharomyces cerevisiae ) NPCs are made up of at least 456 polypeptide chains of ~30 distinct sequences. Many of these components (nucleoporins, Nups) share similar structural motifs and form stable subcomplexes. We have determined a high-resolution crystal structure of the C-terminal domain of yeast Nup133 (ScNup133), a component of the heptameric Nup84 subcomplex. Expression tests yielded ScNup133(944-1157) that produced crystals diffracting to 1.9Å resolution. ScNup133(944-1157) adopts essentially an all α-helical fold, with a short two stranded β-sheet at the C-terminus. The 11 α-helices of ScNup133(944-1157) form a compact fold. In contrast, the previously determined structure of human Nup133(934-1156) bound to a fragment of human Nup107 has its constituent α-helices are arranged in two globular blocks. These differences may reflect structural divergence among homologous nucleoporins.
Organizational Affiliation:
New York SGX Research Center for Structural Genomics, Eli Lilly and Company, Lilly Biotechnology Center, San Diego, California 92121, USA. spartha2@gmail.com