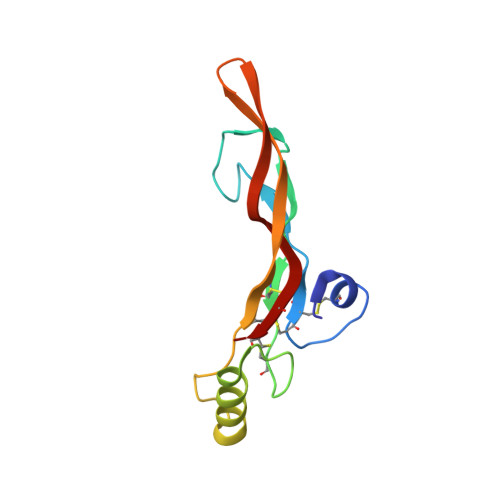
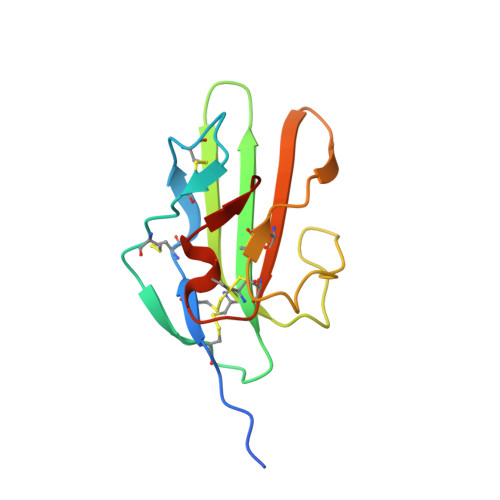
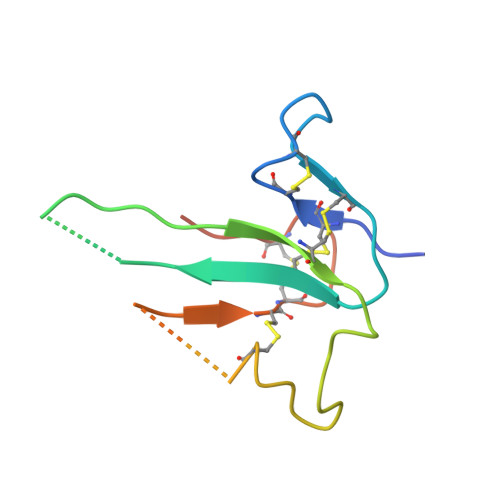
Ternary complex of transforming growth factor-beta1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily.
Radaev, S., Zou, Z., Huang, T., Lafer, E.M., Hinck, A.P., Sun, P.D.(2010) J Biol Chem 285: 14806-14814
- PubMed: 20207738
- DOI: https://doi.org/10.1074/jbc.M109.079921
- Primary Citation of Related Structures:
3KFD - PubMed Abstract:
Transforming growth factor (TGF)-beta1, -beta2, and -beta3 are 25-kDa homodimeric polypeptides that play crucial nonoverlapping roles in embryogenesis, tissue development, carcinogenesis, and immune regulation. Here we report the 3.0-A resolution crystal structure of the ternary complex between human TGF-beta1 and the extracellular domains of its type I and type II receptors, TbetaRI and TbetaRII. The TGF-beta1 ternary complex structure is similar to previously reported TGF-beta3 complex except with a 10 degrees rotation in TbetaRI docking orientation. Quantitative binding studies showed distinct kinetics between the receptors and the isoforms of TGF-beta. TbetaRI showed significant binding to TGF-beta2 and TGF-beta3 but not TGF-beta1, and the binding to all three isoforms of TGF-beta was enhanced considerably in the presence of TbetaRII. The preference of TGF-beta2 to TbetaRI suggests a variation in its receptor recruitment in vivo. Although TGF-beta1 and TGF-beta3 bind and assemble their ternary complexes in a similar manner, their structural differences together with differences in the affinities and kinetics of their receptor binding may underlie their unique biological activities. Structural comparisons revealed that the receptor-ligand pairing in the TGF-beta superfamily is dictated by unique insertions, deletions, and disulfide bonds rather than amino acid conservation at the interface. The binding mode of TbetaRII on TGF-beta is unique to TGF-betas, whereas that of type II receptor for bone morphogenetic protein on bone morphogenetic protein appears common to all other cytokines in the superfamily. Further, extensive hydrogen bonds and salt bridges are present at the high affinity cytokine-receptor interfaces, whereas hydrophobic interactions dominate the low affinity receptor-ligand interfaces.
Organizational Affiliation:
Structural Immunology Section, Laboratory of Immunogenetics, NIAID, National Institutes of Health, Rockville, Maryland 20852, USA.