Crystal structure of a plasmid partition protein from borrelia burgdorferi at 2.25A resolution
Seattle Structural Genomics Center for Infectious Disease (SSGCID), Abendroth, J., Edwards, T., Staker, B.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PF-32 protein | 267 | Borreliella burgdorferi B31 | Mutation(s): 0 Gene Names: PF-32, BB_S35 | 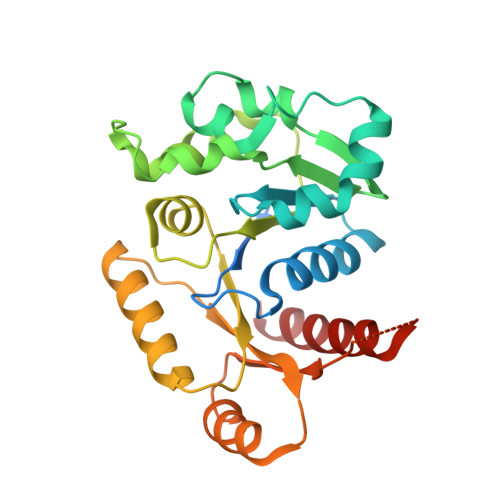 | |
UniProt | |||||
Find proteins for O68233 (Borreliella burgdorferi) Explore O68233 Go to UniProtKB: O68233 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O68233 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | C [auth A], D [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 57 | α = 90 |
| b = 57 | β = 90 |
| c = 343.61 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHASER | phasing |
| REFMAC | refinement |
| XDS | data reduction |
| XSCALE | data scaling |