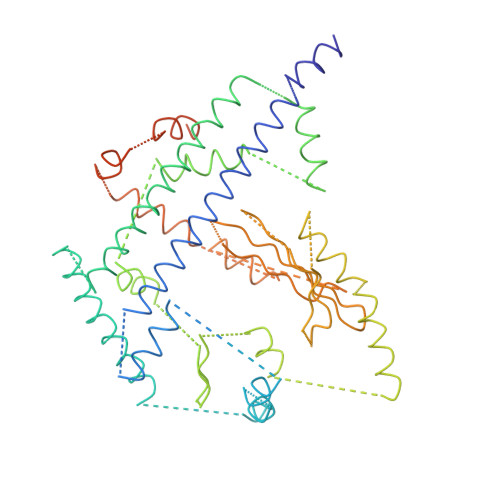
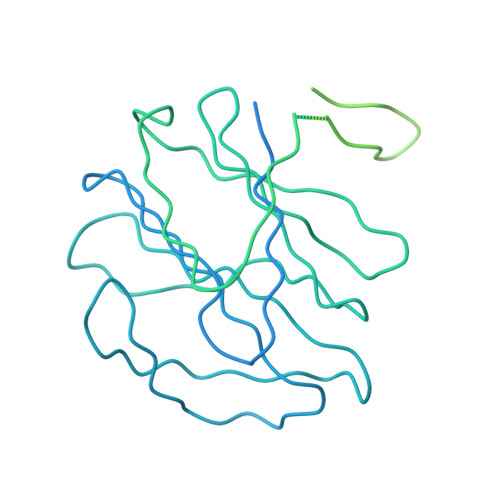
Bluetongue virus coat protein VP2 contains sialic acid-binding domains, and VP5 resembles enveloped virus fusion proteins.
Zhang, X., Boyce, M., Bhattacharya, B., Zhang, X., Schein, S., Roy, P., Zhou, Z.H.(2010) Proc Natl Acad Sci U S A 107: 6292-6297
- PubMed: 20332209
- DOI: https://doi.org/10.1073/pnas.0913403107
- Primary Citation of Related Structures:
3IYK - PubMed Abstract:
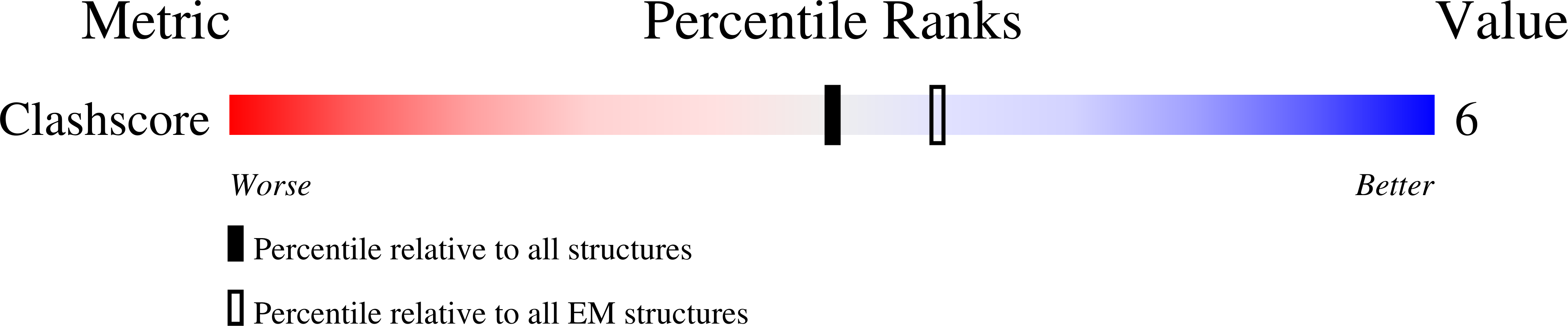
Bluetongue virus (BTV) is transmitted by blood-feeding insects (Culicoides sp.) and causes hemorrhagic diseases in livestock. BTV is a nonenveloped, double-stranded RNA (dsRNA) virus with two capsids: a well-studied, stable core enclosing the dsRNA genome and a highly unstable, poorly studied coat responsible for host cell attachment and entry. Here, based on cryo-electron microscopy (cryoEM), we report a 7-A resolution structure of the infectious BTV virion, including the coat proteins. We show that unlike other dsRNA viruses, the VP2 attachment trimer has a triskelion shape composed of three tip domains branching from a central hub domain. We identify three putative sialic acid-binding pockets in the hub and present supporting biochemical data indicating sugar moiety binding is important for BTV infection. Despite being a nonenveloped virus, the putative VP5 membrane penetration trimer, located slightly inward of the VP2 attachment trimer, has a central coiled-coil alpha-helical bundle, similar to the fusion proteins of many enveloped viruses (e.g., HIV, herpesviruses, vesicular stomatitis virus, and influenza virus). Moreover, mapping of the amino acid sequence of VP5 to the secondary structural elements identified by cryoEM locates 15 amphipathic alpha-helical regions on the external surface of each VP5 trimer. The cryoEM density map also reveals few, weak interactions between the VP5 trimer and both the outer-coat VP2 trimer and the underlying core VP7 trimer, suggesting that the surface of VP5 could unfurl like an umbrella during penetration and shedding of the coat to release the transcriptionally active core particle.
Organizational Affiliation:
Department of Microbiology, Immunology and Molecular Genetics, University of California, Los Angeles, CA 90095-7364, USA.