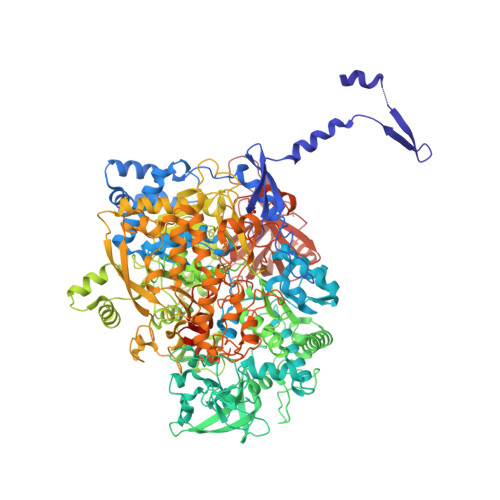
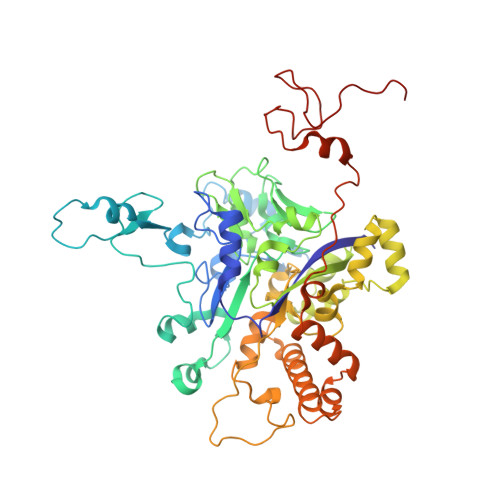
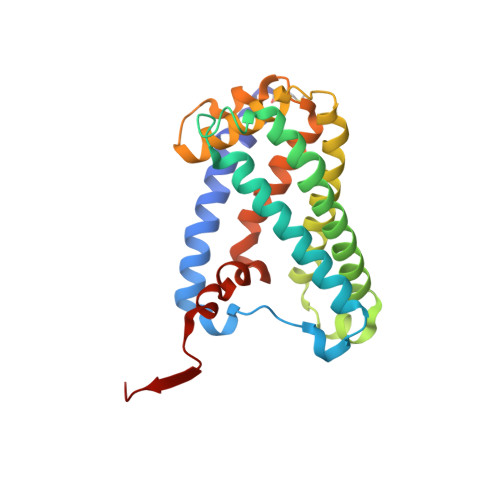
Protein crystallography reveals a role for the FS0 cluster of Escherichia coli nitrate reductase A (NarGHI) in enzyme maturation.
Rothery, R.A., Bertero, M.G., Spreter, T., Bouromand, N., Strynadka, N.C., Weiner, J.H.(2010) J Biol Chem 285: 8801-8807
- PubMed: 20053990
- DOI: https://doi.org/10.1074/jbc.M109.066027
- Primary Citation of Related Structures:
3IR5, 3IR6, 3IR7 - PubMed Abstract:
We have used site-directed mutagenesis, EPR spectroscopy, redox potentiometry, and protein crystallography to monitor assembly of the FS0 [4Fe-4S] cluster and molybdo-bis(pyranopterin guanine dinucleotide) cofactor (Mo-bisPGD) of the Escherichia coli nitrate reductase A (NarGHI) catalytic subunit (NarG). Cys and Ser mutants of NarG-His(49) both lack catalytic activity, with only the former assembling FS0 and Mo-bisPGD. Importantly, both prosthetic groups are absent in the NarG-H49S mutant. EPR spectroscopy of the Cys mutant reveals that the E(m) value of the FS0 cluster is decreased by at least 500 mV, preventing its participation in electron transfer to the Mo-bisPGD cofactor. To demonstrate that decreasing the FS0 cluster E(m) results in decreased enzyme activity, we mutated a critical Arg residue (NarG-Arg(94)) in the vicinity of FS0 to a Ser residue. In this case, the E(m) of FS0 is decreased by 115 mV, with a concomitant decrease in enzyme turnover to approximately 30% of the wild type. Analysis of the structure of the NarG-H49S mutant reveals two important aspects of NarGHI maturation: (i) apomolybdo-NarGHI is able to bind GDP moieties at their respective P and Q sites in the absence of the Mo-bisPGD cofactor, and (ii) a critical segment of residues in NarG, (49)HGVNCTG(55), must be correctly positioned to ensure holoenzyme maturation.
Organizational Affiliation:
Department of Biochemistry, University of Alberta, Edmonton, Alberta T6G 2H7, Canada.