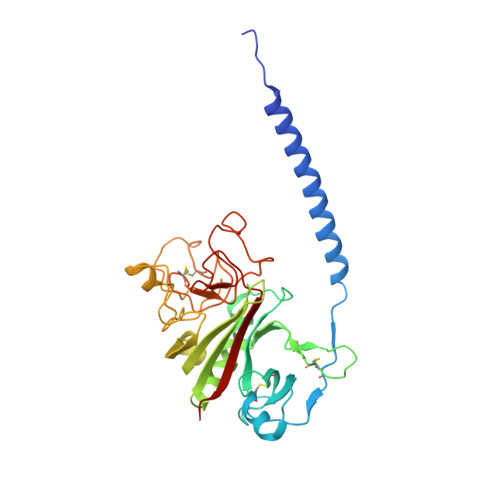
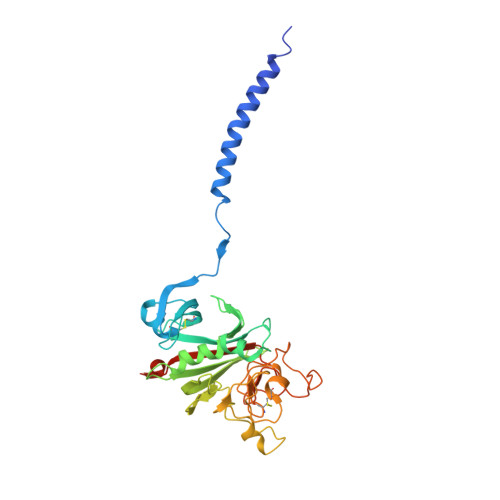
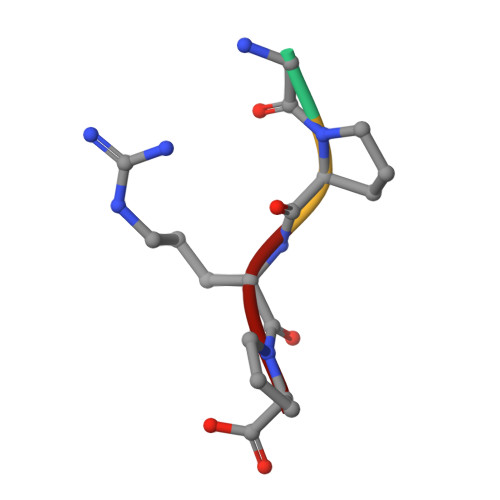
Impaired protofibril formation in fibrinogen gammaN308K is due to altered D:D and "A:a" interactions.
Bowley, S.R., Okumura, N., Lord, S.T.(2009) Biochemistry 48: 8656-8663
- PubMed: 19650644
- DOI: https://doi.org/10.1021/bi900239b
- Primary Citation of Related Structures:
3HUS - PubMed Abstract:
"A:a" knob-hole interactions and D:D interfacial interactions are important for fibrin polymerization. Previous studies with recombinant gammaN308K fibrinogen, a substitution at the D:D interface, showed impaired polymerization. We examined the molecular basis for this loss of function by solving the crystal structure of gammaN308K fragment D. In contrast to previous fragment D crystals, the gammaN308K crystals belonged to a tetragonal space group with an unusually long unit cell (a = b = 95 A, c = 448.3 A). Alignment of the normal and gammaN308K structures showed the global structure of the variant was not changed and the knob "A" peptide GPRP was bound as usual to hole "a". The substitution introduced an elongated positively charged patch in the D:D region. The structure showed novel, symmetric D:D crystal contacts between gammaN308K molecules, indicating the normal asymmetric D:D interface in fibrin would be unstable in this variant. We examined GPRP binding to gammaN308K in solution by plasmin protection assay. The results showed weaker peptide binding, suggesting that "A:a" interactions were altered. We examined fibrin network structures by scanning electron microscopy and found the variant fibers were thicker and more heterogeneous than normal fibers. Considered together, our structural and biochemical studies indicate both "A:a" and D:D interactions are weaker. We conclude that stable protofibrils cannot assemble from gammaN308K monomers, leading to impaired polymerization.
Organizational Affiliation:
Department of Chemistry, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, USA.