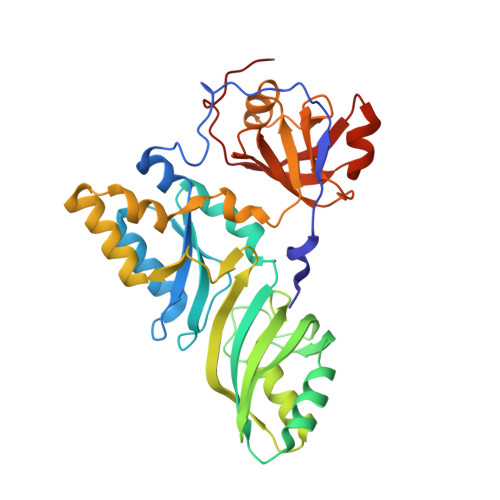
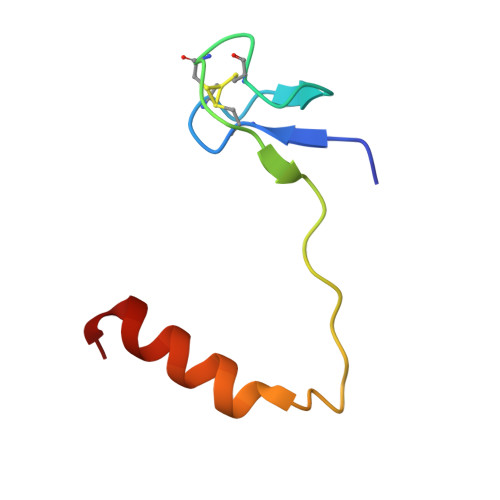
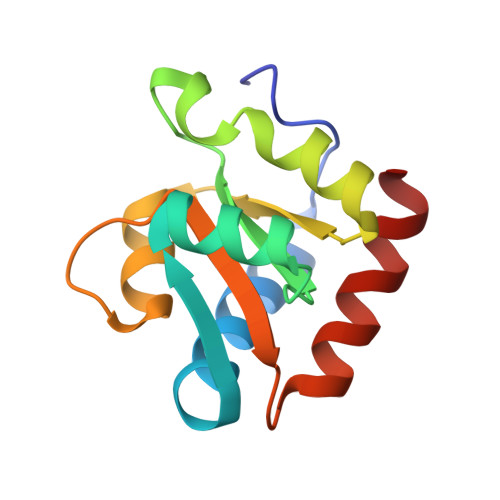
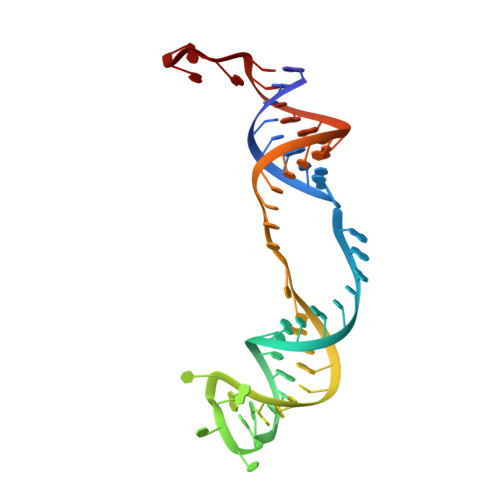
Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA
Liang, B., Zhou, J., Kahen, E., Terns, R.M., Terns, M.P., Li, H.(2009) Nat Struct Mol Biol 16: 740-746
- PubMed: 19478803
- DOI: https://doi.org/10.1038/nsmb.1624
- Primary Citation of Related Structures:
3HJW, 3HJY - PubMed Abstract:
Box H/ACA small nucleolar and Cajal body ribonucleoprotein particles comprise the most complex pseudouridine synthases and are essential for ribosome and spliceosome maturation. The multistep and multicomponent-mediated enzyme mechanism remains only partially understood. Here we report a crystal structure at 2.35 A of a substrate-bound functional archaeal enzyme containing three of the four proteins, Cbf5, Nop10 and L7Ae, and a box H/ACA RNA that reveals detailed information about the protein-only active site. The substrate RNA, containing 5-fluorouridine at the modification position, is fully docked and catalytically rearranged by the enzyme in a manner similar to that seen in two stand-alone pseudouridine synthases. Structural analysis provides a mechanism for plasticity in the diversity of guide RNA sequences used and identifies a substrate-anchoring loop of Cbf5 that also interacts with Gar1 in unliganded structures. Activity analyses of mutated proteins and RNAs support the structural findings and further suggest a role of the Cbf5 loop in regulation of enzyme activity.
Organizational Affiliation:
Institute of Molecular Biophysics, Tallahassee, Florida, USA.