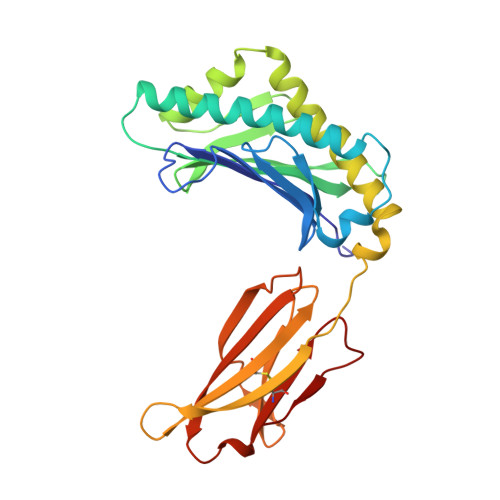
Structural basis of pH-dependent antibody binding by the neonatal Fc receptor.
Vaughn, D.E., Bjorkman, P.J.(1998) Structure 6: 63-73
- PubMed: 9493268
- DOI: https://doi.org/10.1016/s0969-2126(98)00008-2
- Primary Citation of Related Structures:
3FRU - PubMed Abstract:
The neonatal Fc receptor (FcRn) mediates the transcytosis of maternal immunoglobulin G (IgG) across fetal and/or neonatal tissues for the acquisition of passive immunity. In adults, FcRn is involved in the maintenance of high serum IgG levels. Both processes are mediated by pH-dependent IgG binding to FcRn-FcRn binds to IgG with nanomolar affinity at pH 6, but shows no detectable binding at pH 7.5. At pH 6, FcRn is more thermally stable and the dissociation rate of its light chain is an order of magnitude slower than at pH 8.0. Comparison of the structures of FcRn at pH 6.5 and pH 8 allows an analysis of the structural basis for the receptor's pH-dependent ligand binding and stability.
Organizational Affiliation:
Division of Biology, California Institute of Technology, Pasadena 91125, USA.