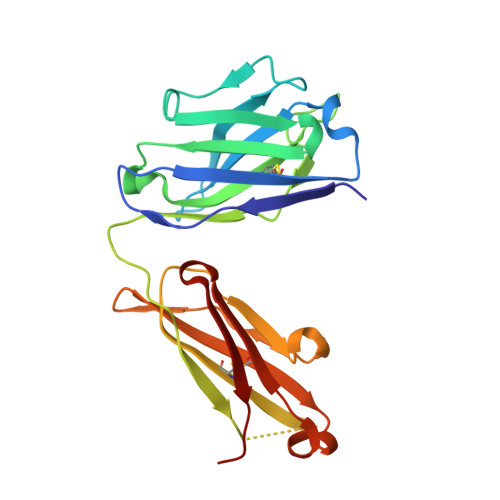
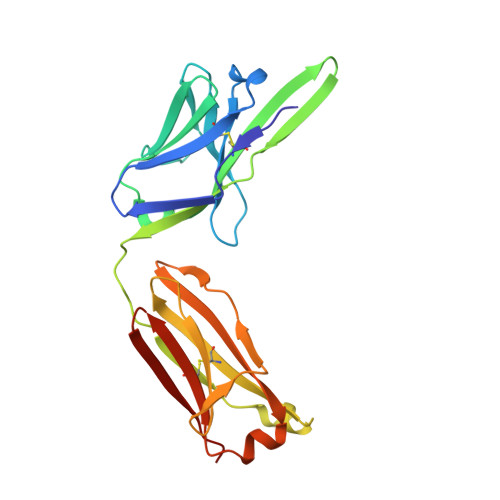
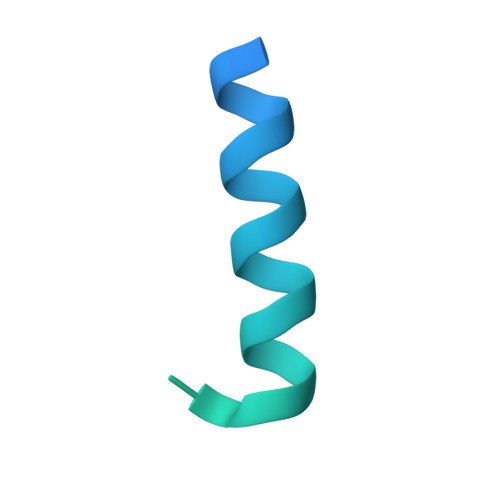
Structural basis for antibody discrimination between two hormones that recognize the parathyroid hormone receptor
McKinstry, W.J., Polekhina, G., Diefenbach-Jagger, H., Ho, P.W.M., Sato, K., Onuma, E., Gillespie, M.T., Martin, T.J., Parker, M.W.(2009) J Biol Chem 284: 15557-15563
- PubMed: 19346515
- DOI: https://doi.org/10.1074/jbc.M900044200
- Primary Citation of Related Structures:
3FFD - PubMed Abstract:
Parathyroid hormone-related protein (PTHrP) plays a vital role in the embryonic development of the skeleton and other tissues. When it is produced in excess by cancers it can cause hypercalcemia, and its local production by breast cancer cells has been implicated in the pathogenesis of bone metastasis formation in that disease. Antibodies have been developed that neutralize the action of PTHrP through its receptor, parathyroid hormone receptor 1, without influencing parathyroid hormone action through the same receptor. Such neutralizing antibodies against PTHrP are therapeutically effective in animal models of the humoral hypercalcemia of malignancy and of bone metastasis formation. We have determined the crystal structure of the complex between PTHrP (residues 1-108) and a neutralizing monoclonal anti-PTHrP antibody that reveals the only point of contact is an alpha-helical structure extending from residues 14-29. Another striking feature is that the same residues that interact with the antibody also interact with parathyroid hormone receptor 1, showing that the antibody and the receptor binding site on the hormone closely overlap. The structure explains how the antibody discriminates between the two hormones and provides information that could be used in the development of novel agonists and antagonists of their common receptor.
Organizational Affiliation:
Biota Structural Biology Laboratory, St. Vincent's Institute of Medical Research, Fitzroy, Victoria 3065, Australia.