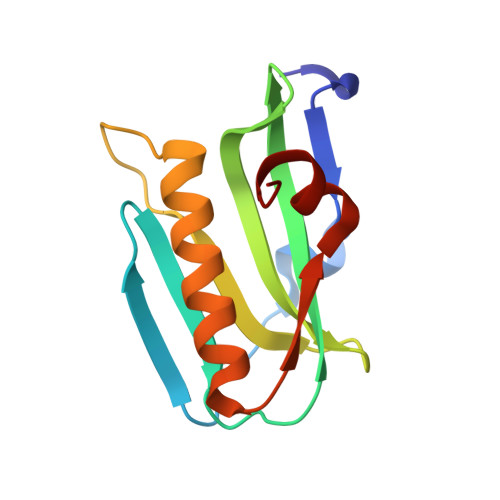
Structure of NS1A effector domain from the influenza A/Udorn/72 virus.
Xia, S., Monzingo, A.F., Robertus, J.D.(2009) Acta Crystallogr D Biol Crystallogr 65: 11-17
- PubMed: 19153461
- DOI: https://doi.org/10.1107/S0907444908032186
- Primary Citation of Related Structures:
3EE8, 3EE9 - PubMed Abstract:
The nonstructural protein NS1A from influenza virus is a multifunctional virulence factor and a potent inhibitor of host immunity. It has two functional domains: an N-terminal 73-amino-acid RNA-binding domain and a C-terminal effector domain. Here, the crystallographic structure of the NS1A effector domain of influenza A/Udorn/72 virus is presented. Structure comparison with the NS1 effector domain from mouse-adapted influenza A/Puerto Rico/8/34 (PR8) virus strain reveals a similar monomer conformation but a different dimer interface. Further analysis and evaluation shows that the dimer interface observed in the structure of the PR8 NS1 effector domain is likely to be a crystallographic packing effect. A hypothetical model of the intact NS1 dimer is presented.
Organizational Affiliation:
Institute for Cellular and Molecular Biology, Department of Chemistry and Biochemistry, University of Texas, 1 University Station A5300, Austin, TX 78712, USA.