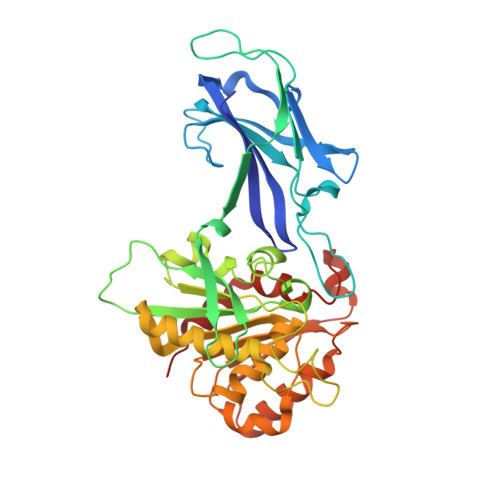
Crystal structure and biochemical properties of a novel thermostable esterase containing an immunoglobulin-like domain.
Levisson, M., Sun, L., Hendriks, S., Swinkels, P., Akveld, T., Bultema, J.B., Barendregt, A., van den Heuvel, R.H.H., Dijkstra, B.W., van der Oost, J., Kengen, S.W.M.(2009) J Mol Biol 385: 949-962
- PubMed: 19013466
- DOI: https://doi.org/10.1016/j.jmb.2008.10.075
- Primary Citation of Related Structures:
3DOH, 3DOI - PubMed Abstract:
Comparative analysis of the genome of the hyperthermophilic bacterium Thermotoga maritima revealed a hypothetical protein (EstA) with typical esterase features. The EstA protein was functionally produced in Escherichia coli and purified to homogeneity. It indeed displayed esterase activity with optima at or above 95 degrees C and at pH 8.5, with a preference for esters with short acyl chains (C2-C10). Its 2.6-A-resolution crystal structure revealed a classical alpha/beta hydrolase domain with a catalytic triad consisting of a serine, an aspartate, and a histidine. EstA is irreversibly inhibited by the organophosphate paraoxon. A 3.0-A-resolution structure confirmed that this inhibitor binds covalently to the catalytic serine residue of EstA. Remarkably, the structure also revealed the presence of an N-terminal immunoglobulin (Ig)-like domain, which is unprecedented among esterases. EstA forms a hexamer both in the crystal and in solution. Electron microscopy showed that the hexamer in solution is identical with the hexamer in the crystal, which is formed by two trimers, with the N-terminal domains facing each other. Mutational studies confirmed that residues Phe89, Phe112, Phe116, Phe246, and Trp377 affect enzyme activity. A truncated mutant of EstA, in which the Ig-like domain was removed, showed only 5% of wild-type activity, had lower thermostability, and failed to form hexamers. These data suggest that the Ig-like domain plays an important role in the enzyme multimerization and activity of EstA.
Organizational Affiliation:
Laboratory of Microbiology, Department of Agrotechnology and Food Sciences, Wageningen University, Dreijenplein 10, 6703 HB Wageningen, The Netherlands. Mark.Levisson@wur.nl