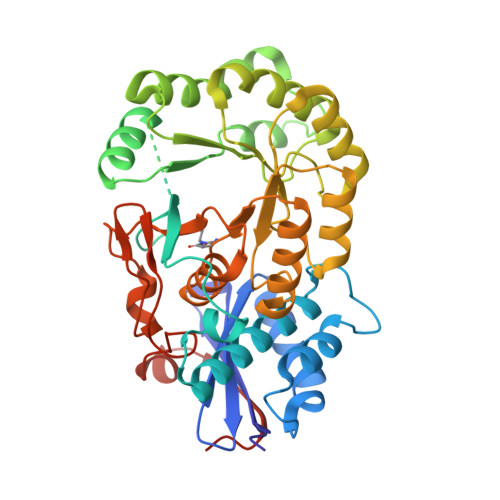
Crystal structure of an enolase protein from the environmental genome shotgun sequencing of the Sargasso Sea
Bonanno, J.B., Freeman, J., Bain, K.T., Zhang, F., Ozyurt, S., Smith, D., Wasserman, S., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.