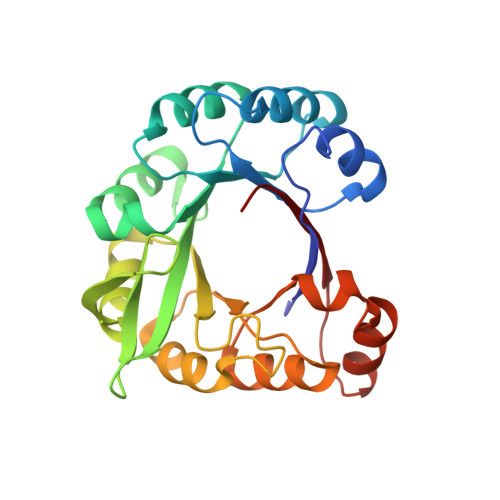
A beta alpha-barrel built by the combination of fragments from different folds.
Bharat, T.A., Eisenbeis, S., Zeth, K., Hocker, B.(2008) Proc Natl Acad Sci U S A 105: 9942-9947
- PubMed: 18632584
- DOI: https://doi.org/10.1073/pnas.0802202105
- Primary Citation of Related Structures:
3CWO - PubMed Abstract:
Combinatorial assembly of protein domains plays an important role in the evolution of proteins. There is also evidence that protein domains have come together from stable subdomains. This concept of modular assembly could be used to construct new well folded proteins from stable protein fragments. Here, we report the construction of a chimeric protein from parts of a (betaalpha)(8)-barrel enzyme from histidine biosynthesis pathway (HisF) and a protein of the (betaalpha)(5)-flavodoxin-like fold (CheY) from Thermotoga maritima that share a high structural similarity. We expected this construct to fold into a full (betaalpha)(8)-barrel. Our results show that the chimeric protein is a stable monomer that unfolds with high cooperativity. Its three-dimensional structure, which was solved to 3.1 A resolution by x-ray crystallography, confirms a barrel-like fold in which the overall structures of the parent proteins are highly conserved. The structure further reveals a ninth strand in the barrel, which is formed by residues from the HisF C terminus and an attached tag. This strand invades between beta-strand 1 and 2 of the CheY part closing a gap in the structure that might be due to a suboptimal fit between the fragments. Thus, by a combination of parts from two different folds and a small arbitrary fragment, we created a well folded and stable protein.
Organizational Affiliation:
Max Planck Institute for Developmental Biology, Spemannstrasse 35, 72076 Tübingen, Germany.