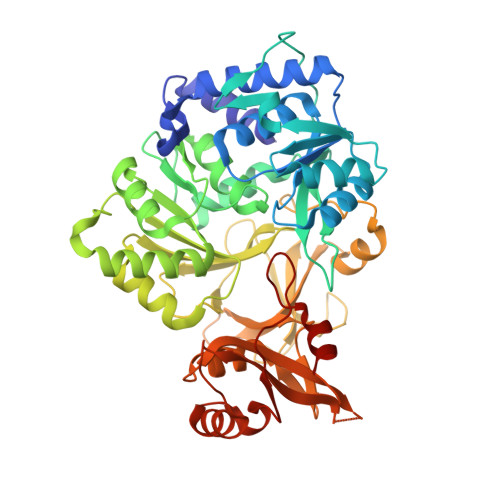
Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Reger, A.S., Wu, R., Dunaway-Mariano, D., Gulick, A.M.(2008) Biochemistry 47: 8016-8025
- PubMed: 18620418
- DOI: https://doi.org/10.1021/bi800696y
- Primary Citation of Related Structures:
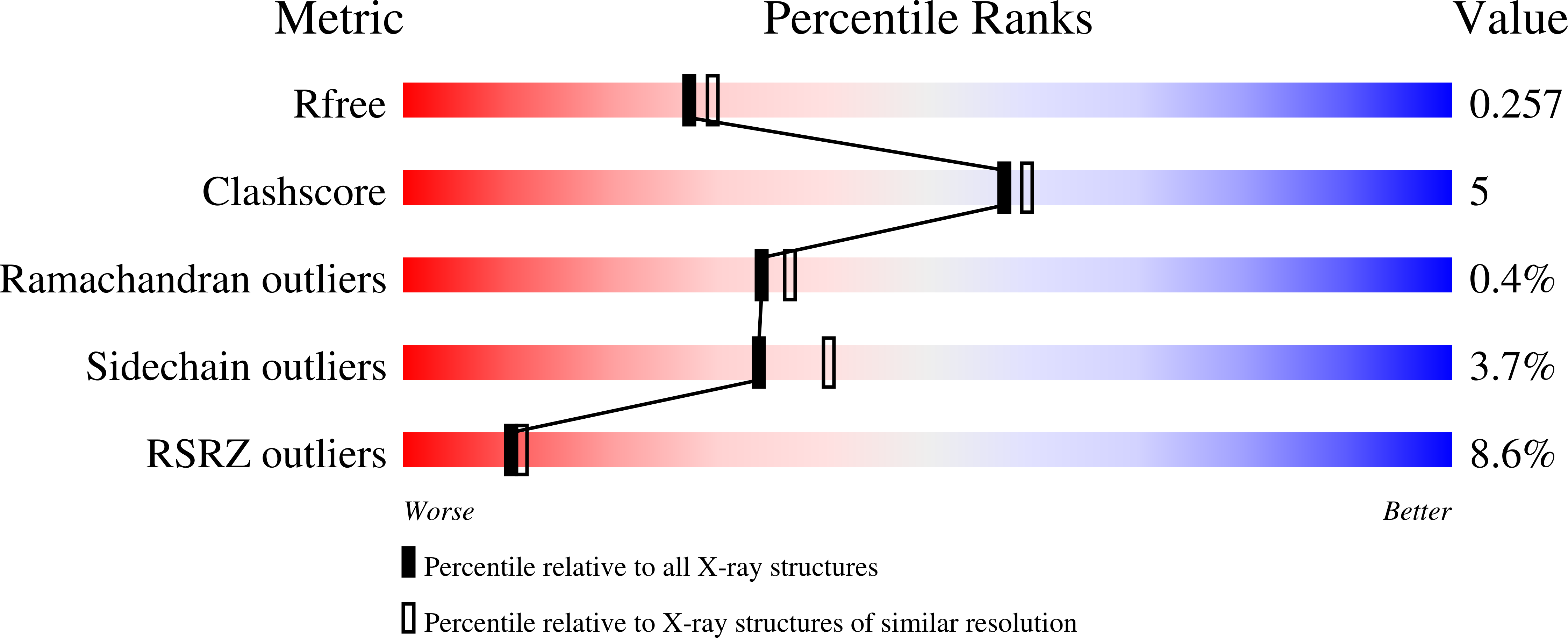
3CW8, 3CW9 - PubMed Abstract:
Members of the adenylate-forming family of enzymes play a role in the metabolism of halogenated aromatics and of short, medium, and long chain fatty acids, as well as in the biosynthesis of menaquinone, peptide antibiotics, and peptide siderophores. This family includes a subfamily of acyl- and aryl-CoA ligases that catalyze thioester synthesis through two half-reactions. A carboxylate substrate first reacts with ATP to form an acyl-adenylate. Subsequent to the release of the product PP i, the enzyme binds CoA, which attacks the activated acyl group to displace AMP. Structural and functional studies on different family members suggest that these enzymes alternate between two conformations during catalysis of the two half-reactions. Specifically, after the initial adenylation step, the C-terminal domain rotates by approximately 140 degrees to adopt a second conformation for thioester formation. Previously, we determined the structure of 4-chlorobenzoate:CoA ligase (CBL) in the adenylate forming conformation bound to 4-chlorobenzoate. We have determined two new crystal structures. We have determined the structure of CBL in the original adenylate-forming conformation, bound to the adenylate intermediate. Additionally, we have used a novel product analogue, 4-chlorophenacyl-CoA, to trap the enzyme in the thioester-forming conformation and determined this structure in a new crystal form. This work identifies a novel binding pocket for the CoA nucleotide. The structures presented herein provide the foundation for biochemical analyses presented in the accompanying manuscript in this issue [Wu et al. (2008) Biochemistry 47, 8026-8039]. The complete characterization of this enzyme allows us to provide an explanation for the use of the domain alternation strategy by these enzymes.
Organizational Affiliation:
Hauptman-Woodward Medical Research Institute, Buffalo, New York 14203, USA.