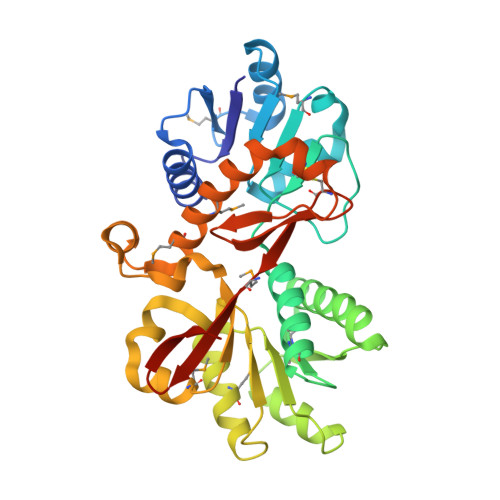
Structure of YraM, a protein essential for growth of Haemophilus influenzae.
Vijayalakshmi, J., Akerley, B.J., Saper, M.A.(2008) Proteins 73: 204-217
- PubMed: 18412262
- DOI: https://doi.org/10.1002/prot.22033
- Primary Citation of Related Structures:
3CKM - PubMed Abstract:
Nontypeable Haemophilus influenzae is an obligate human parasite that often causes middle ear infections in children and exacerbates chronic obstructive pulmonary disorder, the fourth leading cause of death in the United States. There are no effective vaccines available for this strain. The lipoprotein YraM (gene HI1655) was identified as essential for the growth and viability of H. influenzae but its function is unknown. Sequence comparisons showed that YraM is a fusion of two protein modules. We grew crystals of the carboxyl-terminal module of YraM comprising residues 257-573 (YraM-C), phased the diffraction data by the multiwavelength anomalous diffraction technique, and refined the model to a crystallographic R-factor of 0.16 (R(free) = 0.19) with data to 1.35 A resolution. The two-domain structure of YraM-C adopts a fold similar to that observed for the open, unliganded forms of several periplasmic binding proteins (PBPs) involved in bacterial active transport. Sequence alignments of YraM homologues from other Gram-negative species showed that the most conserved residues of YraM-C cluster between the two domains in the location where other PBPs bind their cognate ligand. Modeling of YraM-C into a closed conformation similar to the leucine-bound form of the Leu/Ile/Val-binding protein (LIVBP) shows a putative binding pocket larger than the leucine-binding site in LIVBP. The pocket has both polar and nonpolar surfaces, with the latter located in the same area where a leucine side chain binds to LIVBP. We discuss possible biological functions of YraM considering its predicted location in the outer membrane, a novel place for such a binding protein.
Organizational Affiliation:
Biophysics, University of Michigan, Ann Arbor, Michigan 48109-1055, USA.