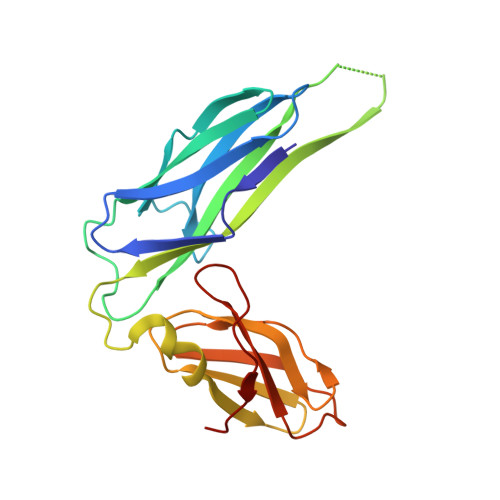
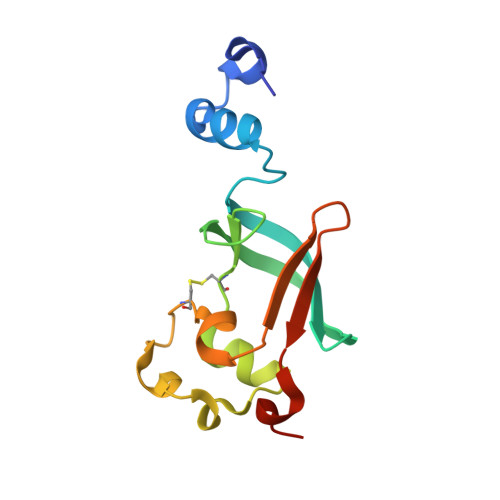
Crystal structure of the ternary FimC-FimF(t)-FimD(N) complex indicates conserved pilus chaperone-subunit complex recognition by the usher FimD
Eidam, O., Dworkowski, F.S., Glockshuber, R., Grutter, M.G., Capitani, G.(2008) FEBS Lett 582: 651-655
- PubMed: 18242189
- DOI: https://doi.org/10.1016/j.febslet.2008.01.030
- Primary Citation of Related Structures:
3BWU - PubMed Abstract:
Type 1 pili, anchored to the outer membrane protein FimD, enable uropathogenic Escherichia coli to attach to host cells. During pilus biogenesis, the N-terminal periplasmic domain of FimD (FimD(N)) binds complexes between the chaperone FimC and pilus subunits via its partly disordered N-terminal segment, as recently shown for the FimC-FimH(P)-FimD(N) ternary complex. We report the structure of a new ternary complex (FimC-FimF(t)-FimD(N)) with the subunit FimF(t) instead of FimH(p). FimD(N) recognizes FimC-FimF(t) and FimC-FimH(P) very similarly, predominantly through hydrophobic interactions. The conserved binding mode at a "hot spot" on the chaperone surface could guide the design of pilus assembly inhibitors.
Organizational Affiliation:
University of Zurich, Department of Biochemistry, Winterthurerstrasse 190, Zurich, Switzerland.