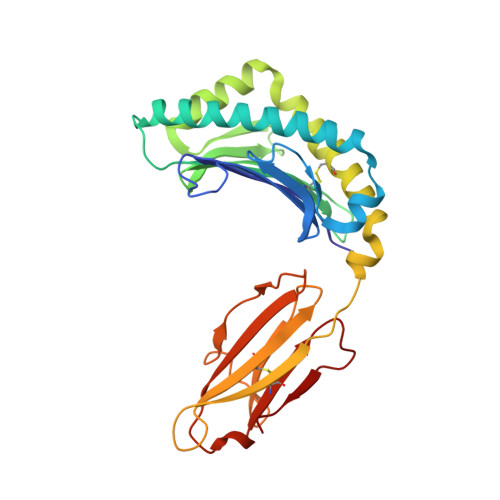
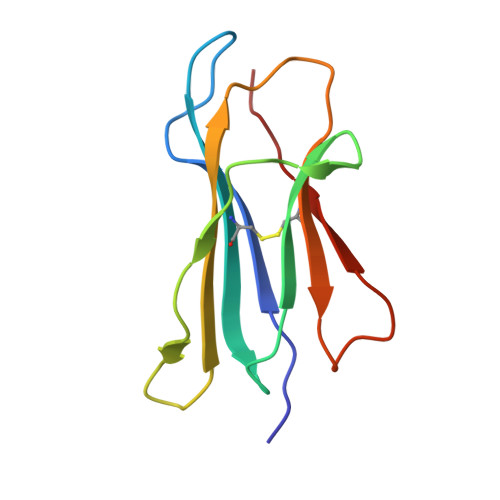
Structures of an MHC class I molecule from b21 chickens illustrate promiscuous Peptide binding
Koch, M., Camp, S., Collen, T., Avila, D., Salomonsen, J., Wallny, H.-J., van Hateren, A., Hunt, L., Jacob, J.P., Johnston, F., Marston, D.A., Shaw, I., Dunbar, P.R., Cerundolo, V., Jones, E.Y., Kaufman, J.(2007) Immunity 27: 885-899
- PubMed: 18083574
- DOI: https://doi.org/10.1016/j.immuni.2007.11.007
- Primary Citation of Related Structures:
3BEV, 3BEW - PubMed Abstract:
Little is known about the structure of major histocompatibility complex (MHC) molecules outside of mammals. Only one class I molecule in the chicken MHC is highly expressed, leading to strong genetic associations with infectious pathogens. Here, we report two structures of the MHC class I molecule BF2*2101 from the B21 haplotype, which is known to confer resistance to Marek's disease caused by an oncogenic herpesvirus. The binding groove has an unusually large central cavity, which confers substantial conformational flexibility to the crucial residue Arg9, allowing remodeling of key peptide-binding sites. The coupled variation of anchor residues from the peptide, utilizing a charge-transfer system unprecedented in MHC molecules, allows peptides with conspicuously different sequences to be bound. This promiscuous binding extends our understanding of ways in which MHC class I molecules can present peptides to the immune system and might explain the resistance of the B21 haplotype to Marek's disease.
Organizational Affiliation:
Cancer Research UK Receptor Structure Research Group, The Henry Wellcome Building for Genomic Medicine, Roosevelt Drive, Headington, Oxford OX3 7BN, UK.