Insights into the substrate specificity of a thioesterase Rv0098 of mycobacterium tuberculosis through X-ray crystallographic and molecular dynamics studies.
Maity, K., Bajaj, P., Surolia, N., Surolia, A., Suguna, K.(2012) J Biomol Struct Dyn 29: 973-983
- PubMed: 22292955
- DOI: https://doi.org/10.1080/07391102.2012.10507417
- Primary Citation of Related Structures:
3B18 - PubMed Abstract:
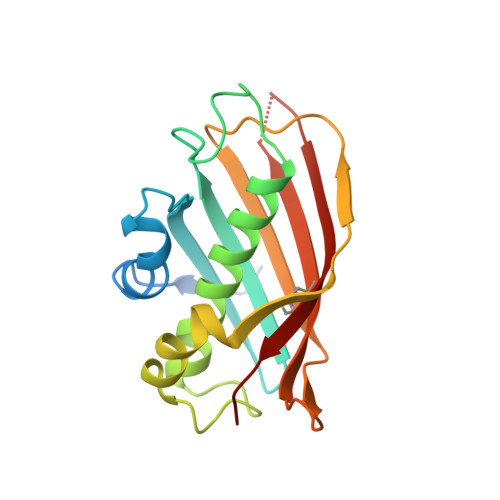
The crystal structure of Rv0098, a long-chain fatty acyl-CoA thioesterase from Mycobacterium tuberculosis with bound dodecanoic acid at the active site provided insights into the mode of substrate binding but did not reveal the structural basis of substrate specificities of varying chain length. Molecular dynamics studies demonstrated that certain residues of the substrate binding tunnel are flexible and thus modulate the length of the tunnel. The flexibility of the loop at the base of the tunnel was also found to be important for determining the length of the tunnel for accommodating appropriate substrates. A combination of crystallographic and molecular dynamics studies thus explained the structural basis of accommodating long chain substrates by Rv0098 of M. tuberculosis.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore - 560 012, India.