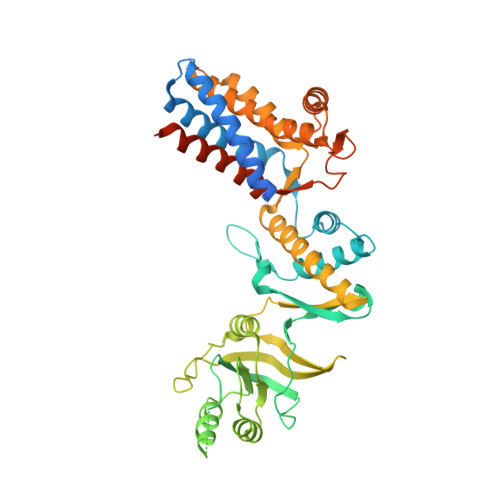
Chaperonins from an Antarctic archaeon are predominantly monomeric: crystal structure of an open state monomer.
Pilak, O., Harrop, S.J., Siddiqui, K.S., Chong, K., De Francisci, D., Burg, D., Williams, T.J., Cavicchioli, R., Curmi, P.M.(2011) Environ Microbiol 13: 2232-2249
- PubMed: 21477108
- DOI: https://doi.org/10.1111/j.1462-2920.2011.02477.x
- Primary Citation of Related Structures:
3AQ1 - PubMed Abstract:
Archaea are abundant in permanently cold environments. The Antarctic methanogen, Methanococcoides burtonii, has proven an excellent model for studying molecular mechanisms of cold adaptation. Methanococcoides burtonii contains three group II chaperonins that diverged prior to its closest orthologues from mesophilic Methanosarcina spp. The relative abundance of the three chaperonins shows little dependence on organism growth temperature, except at the highest temperatures, where the most thermally stable chaperonin increases in abundance. In vitro and in vivo, the M. burtonii chaperonins are predominantly monomeric, with only 23-33% oligomeric, thereby differing from other archaea where an oligomeric ring form is dominant. The crystal structure of an N-terminally truncated chaperonin reveals a monomeric protein with a fully open nucleotide binding site. When compared with closed state group II chaperonin structures, a large-scale ≈ 30° rotation between the equatorial and intermediate domains is observed resulting in an open nucleotide binding site. This is analogous to the transition observed between open and closed states of group I chaperonins but contrasts with recent archaeal group II chaperonin open state ring structures. The predominance of monomeric form and the ability to adopt a fully open nucleotide site appear to be unique features of the M. burtonii group II chaperonins.
Organizational Affiliation:
School of Biotechnology and Biomolecular Sciences, The University of New South Wales, Sydney, New South Wales 2052, Australia.