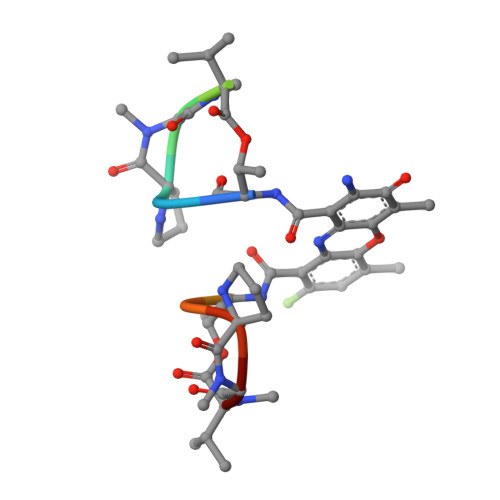
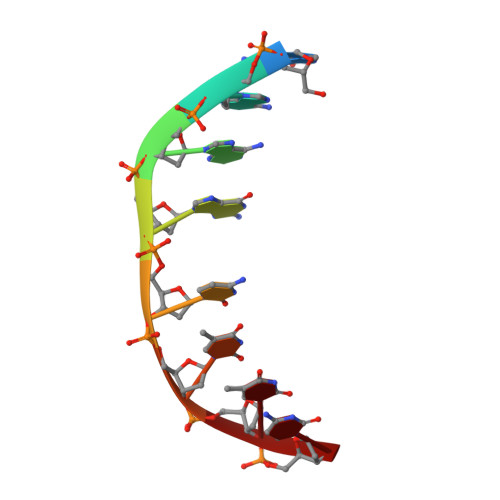
Selectivity of F8-Actinomycin D for RNA:DNA Hybrids and its Anti-Leukemia Activity.
Takusagawa, F., Takusagawa, K.T., Carlson, R.G., Weaver, R.F.(1997) Bioorg Med Chem 5: 1197
- PubMed: 9222513
- DOI: https://doi.org/10.1016/s0968-0896(97)00062-x
- Primary Citation of Related Structures:
316D - PubMed Abstract:
Although many compounds have been found that bind to DNA in various ways and exhibit various biological activities, few compounds that specifically bind to RNA or RNA:DNA hybrids are known, even though such compounds are expected to have important biological properties. For example, one characteristic function of the retroviruses, which is generally not found in eukaryotic cells, is the production of an RNA:DNA hybrid in the viral replication phase. If an agent is designed to bind only to an RNA:DNA hybrid, and not to DNA or to RNA, such an agent might be able to inhibit specifically the RNase H activity of retroviral reverse transcriptase, and therefore suppress viral replication. Actinomycin D is known to bind to double-stranded DNA, but not to RNA, because steric hindrance between the 2-amino group of the phenoxazone ring and the 2'-hydroxyl group of RNA prevents intercalation of the compound. However, if the > C-H moiety at the 8-position of the phenoxazone ring is replaced by a > C-F, a possible hydrogen-bond acceptor, this analogue (8-fluoro-actinomycin D, F8AMD) might be able to bind intercalatively to an RNA:DNA hybrid by forming an additional hydrogen bond between F8 and the 2'-hydroxyl group of the guanosine ribose. To test this hypothesis, the crystal structure of d(GAAGCTTC)2-F8AMD has been determined at 3.0 A resolution. Based on this crystal structure, a model in which F8AMD binds into the hybrid r(GAAGCUUC):d(GAAGCTTC) has been built using molecular mechanics and dynamic methods. These structural studies indicate that F8AMD binds intercalatively to a B-form double-stranded DNA whereas the drug intercalates into an RNA:DNA hybrid taking an A-form conformation. In the RNA:DNA hybrid complex, the F8 atom is located so as to be able to interact to an O2' hydroxyl group with either an O-H...F hydrogen bond or H+...F- electrostatic interaction. This interaction might stabilize the F8AMD molecule in the RNA:DNA hybrid. A binding study indicates that both actinomycin D (AMD) and F8AMD bind intercalatively not only to double-stranded DNAs, but also to RNA:DNA hybrids. Although the overall binding capacity of F8AMD (k = 4.5 x 10(5) M-1) is reduced slightly in comparison with AMD itself (k = 1.8 x 10(6) M-1), F8AMD tends to bind relatively more favorably than AMD to the RNA:DNA hybrids. The drugs' effects on RNA synthesis in HeLa cells indicates that the binding capacities of AMD and F8AMD correlates strongly to their RNA synthesis inhibitory activities. F8AMD required a concentration of 78 nM to inhibit RNA polymerase activity in HeLa cells by 50%, whereas AMD reached the same inhibitory level at 30 nM. Surprisingly, F8AMD exhibits unique selectivity against leukemia cells as does another C8-derivatized AMD analogue, N8AMD. F8AMD inhibits 50% of leukemia cell growth at less than 1.0 nM whereas 10- to 130-fold-higher drug concentrations are required to inhibit the growth of other tumor cell lines by 50%. The GI50 value of F8AMD for leukemia cells is the lowest among the GI50 values for all other AMD derivatives tested. By contrast, AMD is quite potent and kills most cells at less than 50 nM concentration, but it does not show any selectivity for certain cell lines. This indicates that AMD should have very limited use as an antitumor agent. It is difficult to rationalize why F8AMD and N8AMD show such strong selectivity against leukemia cells. However, this study and our previous study (J. Am. Chem. Soc. 1994, 116, 7971) indicated that F8AMD and N8AMD tended to bind more favorably to RNA:DNA hybrids. Thus, the unique antileukemia selectivity shown by F8AMD and N8AMD might be used by the agents binding to RNA:DNA hybrids rather than to double-stranded DNA.
Organizational Affiliation:
Department of Biochemistry, University of Kansas, Lawrence 66045-2106, USA.