Crystal structure and mutational analysis of Ca2+-independent type II antifreeze protein from longsnout poacher, Brachyopsis rostratus
Nishimiya, Y., Kondo, H., Takamichi, M., Sugimoto, H., Suzuki, M., Miura, A., Tsuda, S.(2008) J Mol Biol 382: 734-746
- PubMed: 18674542
- DOI: https://doi.org/10.1016/j.jmb.2008.07.042
- Primary Citation of Related Structures:
2ZIB - PubMed Abstract:
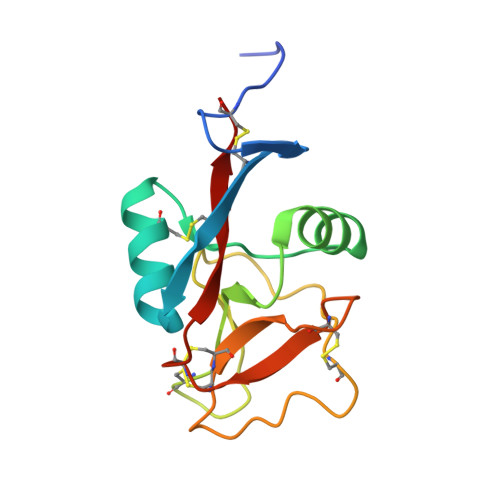
We recently found that longsnout poacher (Brachyosis rostratus) produces a Ca(2+)-independent type II antifreeze protein (lpAFP) and succeeded in expressing recombinant lpAFP using Phichia pastoris. Here, we report, for the first time, the X-ray crystal structure of lpAFP at 1.34 A resolution. The lpAFP structure displayed a relatively planar surface, which encompasses two loop regions (Cys86-Lys89 and Asn91-Cys97) and a short beta-strand (Trp109-Leu112) with three unstructured segments (Gly57-Ile58, Ala103-Ala104, and Pro113-His118). Electrostatic calculation of the protein surface showed that the relatively planar surface was divided roughly into a hydrophobic area (composed of the three unstructured segments lacking secondary structure) and a hydrophilic area (composed of the loops and beta-strand). Site-directed mutation of Ile58 with Phe at the center of the hydrophobic area decreased activity significantly, whereas mutation of Leu112 with Phe at an intermediate area between the hydrophobic and hydrophilic areas retained complete activity. In the hydrophilic area, a peptide-swap mutant in the loops retained 60% activity despite simultaneous mutations of eight residues. We conclude that the epicenter of the ice-binding site of lpAFP is the hydrophobic region, which is centered by Ile58, in the relatively planar surface. We built an ice-binding model for lpAFP on the basis of a lattice match of ice and constrained water oxygen atoms surrounding the hydrophobic area in the lpAFP structure. The model in which lpAFP has been docked to a secondary prism (2-1-10) plane, which is different from the one determined for Ca(2+)-independent type II AFP from sea raven (11-21), appears to explain the results of the mutagenesis analysis.
Organizational Affiliation:
Functional Protein Research Group, Research Institute of Genome-based Biofactory, National Institute of Advanced Industrial Science and Technology, 2-17-2-1 Tsukisamu-Higashi, Toyohira, Sapporo 062-8517, Japan.