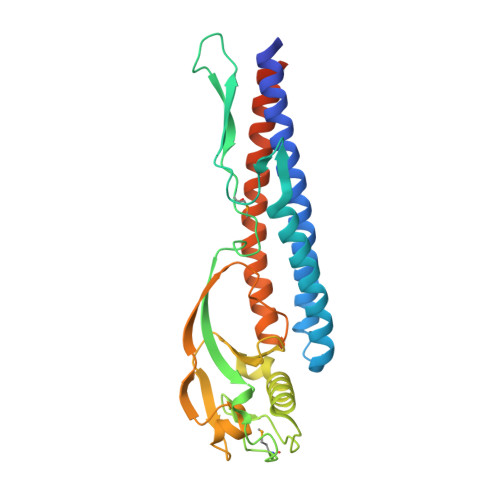
Crystal structure of a novel bacterial cell-surface flagellin binding to a polysaccharide
Maruyama, Y., Momma, M., Mikami, B., Hashimoto, W., Murata, K.(2008) Biochemistry 47: 1393-1402
- PubMed: 18189419
- DOI: https://doi.org/10.1021/bi701872x
- Primary Citation of Related Structures:
2ZBI - PubMed Abstract:
Bacterial flagellins are generally self-assembled into extracellular flagella for cell motility. However, the flagellin homologue p5 is found on the cell surface of Sphingomonas sp. A1 (strain A1) and binds tightly to the alginate polysaccharide. To assimilate alginate, strain A1 forms a mouthlike pit on the cell surface and concentrates the polymer in the pit. p5 is a candidate receptor that recognizes extracellular alginate and controls pit formation. To improve our understanding of the structure and function of p5, we determined the crystal structure of truncated p5 (p5DeltaN53C45) at 2.0 A resolution. This, to our knowledge, is the first structure of flagellin_IN motif-containing flagellin. p5DeltaN53C45 consists of two domains: an alpha-domain rich in alpha-helices that forms the N- and C-terminal regions and a beta-domain rich in beta-strands that constitutes the central region. The alpha-domain is structurally similar to the D1 domain of Salmonella typhimurium flagellin, while the beta-domain is structurally similar to the finger domain of the bacteriophage T4 baseplate protein that is important for intermolecular interactions between baseplate and a long or short tail fiber. Results from the deletion mutant analysis suggest that residues 20-40 and 353-363 are responsible for alginate binding. Truncated N- and C-terminal regions are thought to constitute alpha-helices extending from the alpha-domain. On the basis of the size and surface charge, the cleft in extended alpha-helices is proposed as an alginate binding site of p5. Structural similarity in the beta-domain suggests that the beta-domain is involved in the proper localization and/or orientation of p5 on the cell surface.
Organizational Affiliation:
Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Uji, Kyoto 611-0011, Japan.