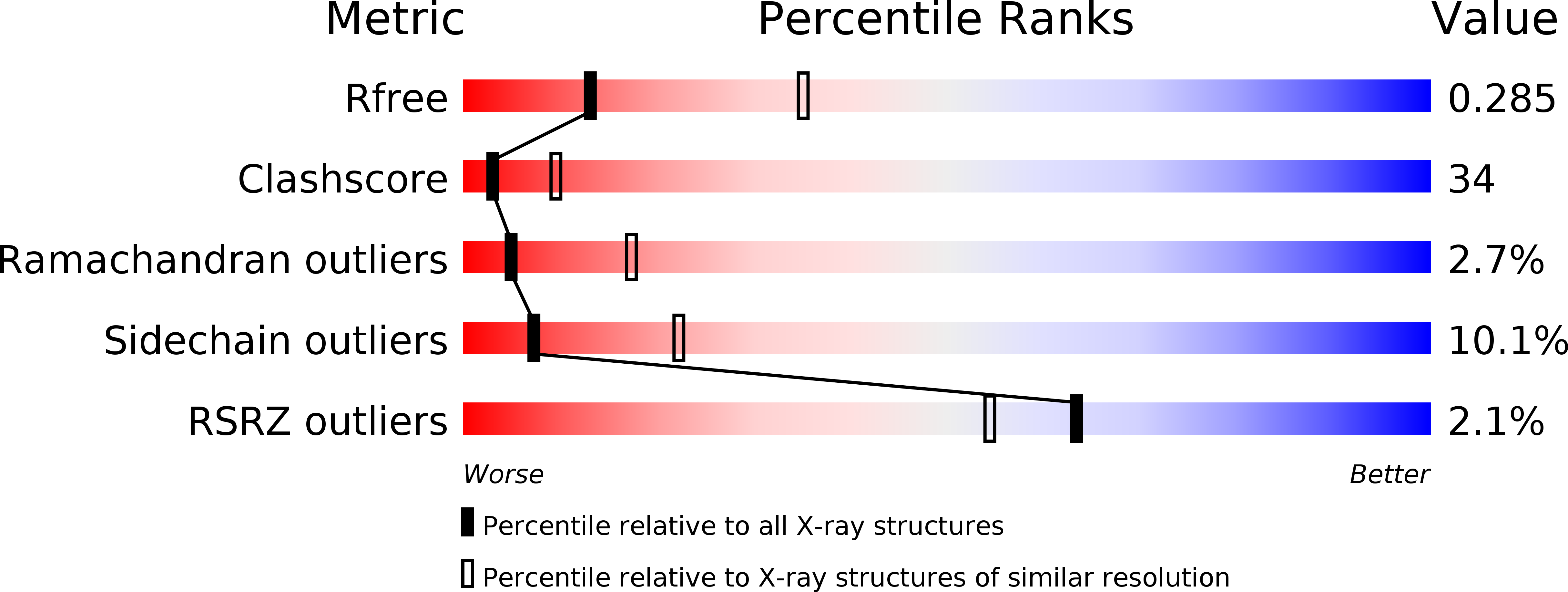
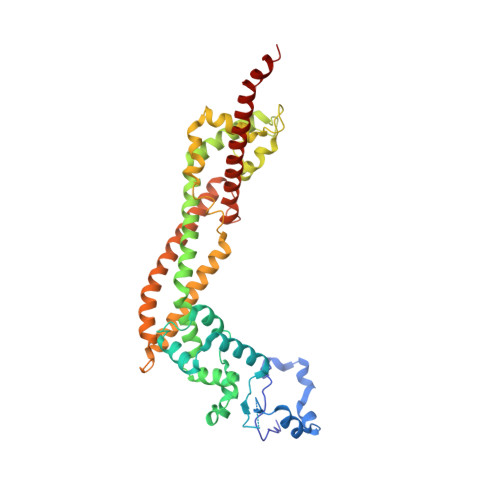
Crystal structure of human DAAM1 formin homology 2 domain
Yamashita, M., Higashi, T., Suetsugu, S., Sato, Y., Ikeda, T., Shirakawa, R., Kita, T., Takenawa, T., Horiuchi, H., Fukai, S., Nureki, O.(2007) Genes Cells 12: 1255-1265
- PubMed: 17986009
- DOI: https://doi.org/10.1111/j.1365-2443.2007.01132.x
- Primary Citation of Related Structures:
2Z6E - PubMed Abstract:
Reorganization of the actin filament is an essential process for cell motility, cell-cell attachment and intracellular transport. Formin proteins promote nucleation and elongation of the actin filament, and thus are key regulators for this process. The formin homology 2 (FH2) domain forms a head-to-tail ring-shaped dimer, and processively moves towards the barbed end. Dishevelled-associated activator of morphogenesis (DAAM) is a Rho-regulated formin implicated in neuronal development. Here, we present the crystal structure of human DAAM1 FH2 dimer at 2.8 A resolution. This is the first dimeric structure of the mammalian formin. The core structure of human DAAM1 is similar to those of mouse mDia1 and yeast Bni1p, whereas the orientations of the FH2 dimeric rings are different between human DAAM1 and yeast Bni1p, despite their similar dimer interactions. This difference supports the previous prediction that the dimer architecture of the formin is highly flexible in the actin-free state. The results of the actin assembly assays using the DAAM1 mutants demonstrated that the length of the linker connecting the N-terminal domain and the core region is crucial for the activity.
Organizational Affiliation:
Department of Biological Information, Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology, 4259 Nagatsuta-cho, Midori-ku, Yokohama-shi, Kanagawa 226-8501, Japan.