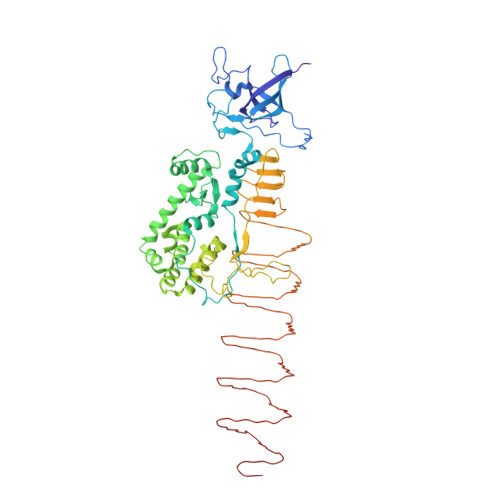
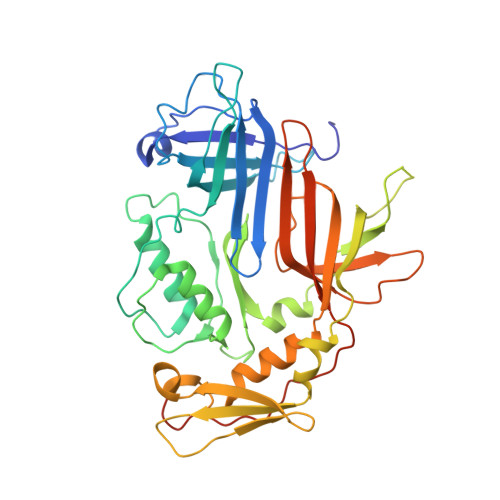
Molecular design of heteroprotein assemblies providing a bionanocup as a chemical reactor.
Koshiyama, T., Yokoi, N., Ueno, T., Kanamaru, S., Nagano, S., Shiro, Y., Arisaka, F., Watanabe, Y.(2008) Small 4: 50-54
- PubMed: 18098245
- DOI: https://doi.org/10.1002/smll.200700855
- Primary Citation of Related Structures:
2Z6B
Organizational Affiliation:
Department of Chemistry, Graduate School of Science, Nagoya University, Furo-cho, Chikusa-ku, Nagoya 464-8602, Japan.