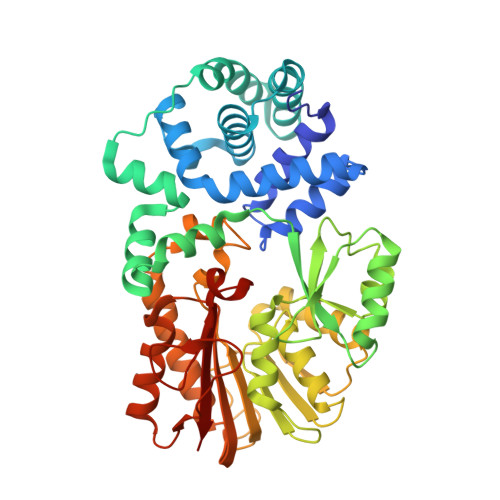
Structure of an archaeal homologue of the bacterial Fmu/RsmB/RrmB rRNA cytosine 5-methyltransferase
Hikida, Y., Kuratani, M., Bessho, Y., Sekine, S., Yokoyama, S.(2010) Acta Crystallogr D Biol Crystallogr 66: 1301-1307
- PubMed: 21123870
- DOI: https://doi.org/10.1107/S0907444910037558
- Primary Citation of Related Structures:
2YXL - PubMed Abstract:
One of the modified nucleosides that frequently occurs in rRNAs and tRNAs is 5-methylcytidine (m⁵C). Escherichia coli Fmu/RsmB/RrmB is an S-adenosyl-L-methionine (AdoMet)-dependent methyltransferase that forms m⁵C967 in 16S rRNA. Fmu/RsmB/RrmB homologues exist not only in bacteria but also in archaea and eukarya and constitute a large orthologous group in the RNA:m⁵C methyltransferase family. In the present study, the crystal structure of a homologue of E. coli Fmu/RsmB/RrmB from the archaeon Pyrococcus horikoshii (PH0851) complexed with an AdoMet analogue was determined at 2.55 Å resolution. The structure and sequence of the C-terminal catalytic domain are highly conserved compared with those of E. coli Fmu/RsmB/RrmB. In contrast, the sequence of the N-terminal domain is negligibly conserved between the bacterial and archaeal subfamilies. Nevertheless, the N-terminal domains of PH0851 and E. coli Fmu/RsmB/RrmB are both α-helical and adopt a similar topology. Next to the AdoMet-binding site, a positively charged cleft is formed between the N- and C-terminal domains. This cleft is conserved in the archaeal PH0851 homologues and seems to be suitable for binding the RNA substrate.
Organizational Affiliation:
Department of Biophysics and Biochemistry, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan.