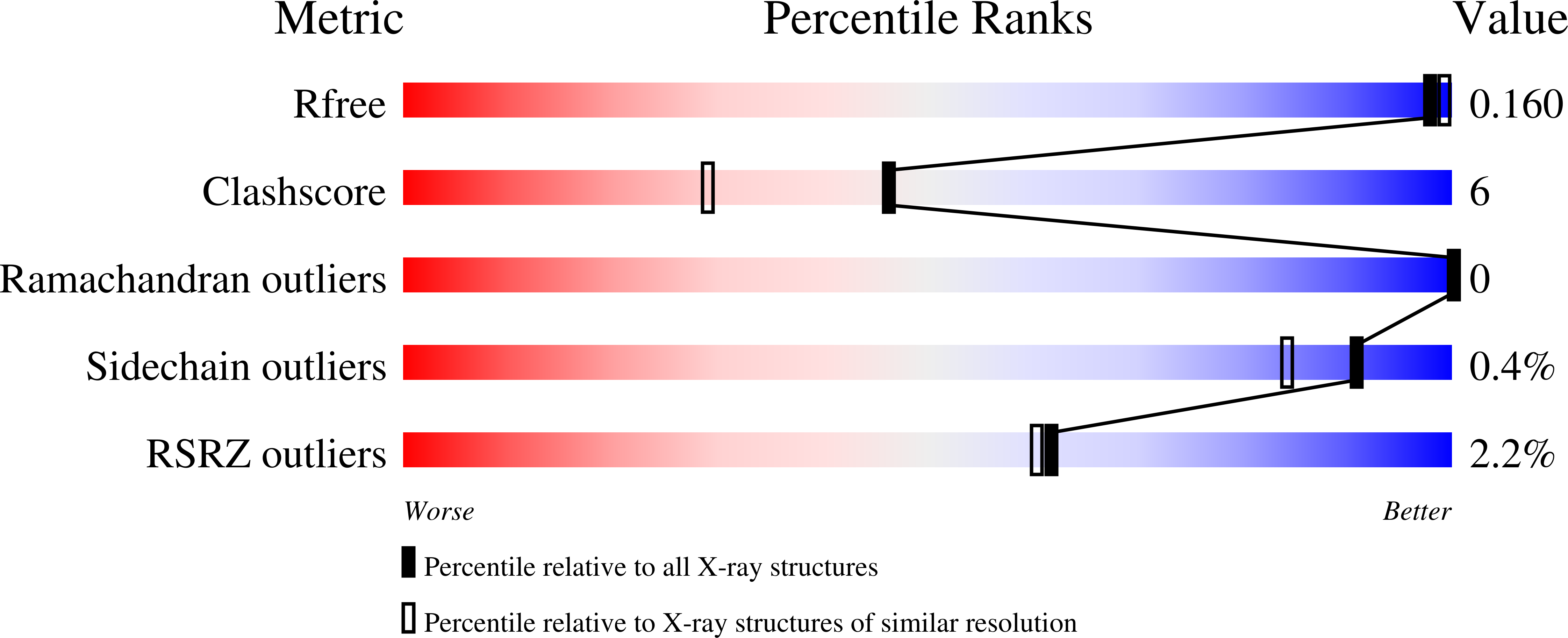
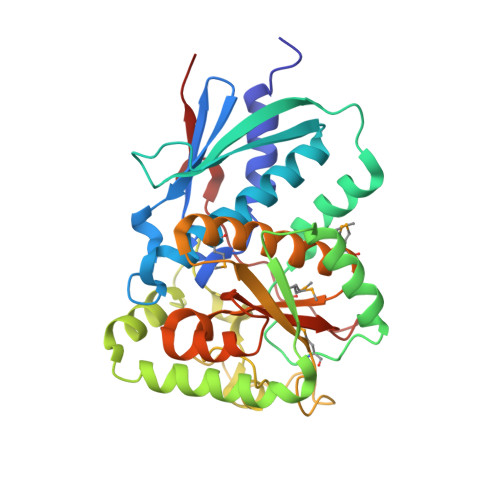
Crystal Structure of Reduced Msacg, a Putative Nitroreductase from Mycobacterium Smegmatis and a Close Homologue of Mycobacterium Tuberculosis Acg.
Chauviac, F.-X., Bommer, M., Yan, J., Parkin, G., Daviter, T., Lowden, P., Raven, E.L., Thalassinos, K., Keep, N.H.(2012) J Biol Chem 287: 44372
- PubMed: 23148223
- DOI: https://doi.org/10.1074/jbc.M112.406264
- Primary Citation of Related Structures:
2YMV - PubMed Abstract:
This paper presents the structure of MsAcg (MSMEG_5246), a Mycobacterium smegmatis homologue of Mycobacterium tuberculosis Acg (Rv2032) in its reduced form at 1.6 Å resolution using x-ray crystallography. Rv2032 is one of the most induced genes under the hypoxic model of tuberculosis dormancy. The Acg family turns out to be unusual flavin mononucleotide (FMN)-binding proteins that have probably arisen by gene duplication and fusion from a classical homodimeric nitroreductase such that the monomeric protein resembles a classical nitroreductase dimer but with one active site deleted and the other active site covered by a unique lid. The FMN cofactor is not reduced by either NADH or NADPH, but the chemically reduced enzyme is capable of reduction of nitro substrates, albeit at no kinetic advantage over free FMN. The reduced enzyme is rapidly oxidized by oxygen but without any evidence for a radical state commonly seen in oxygen-sensitive nitroreductases. The presence of the unique lid domain, the lack of reduction by NAD(P)H, and the slow rate of reaction of the chemically reduced protein raises a possible alternative function of Acg proteins in FMN storage or sequestration from other biochemical pathways as part of the bacteria's adaptation to a dormancy state.
Organizational Affiliation:
Crystallography, Institute for Structural and Molecular Biology, Department of Biological Sciences, Birkbeck, University of London, Malet Street, London, WC1E 7HX, United Kingdom.