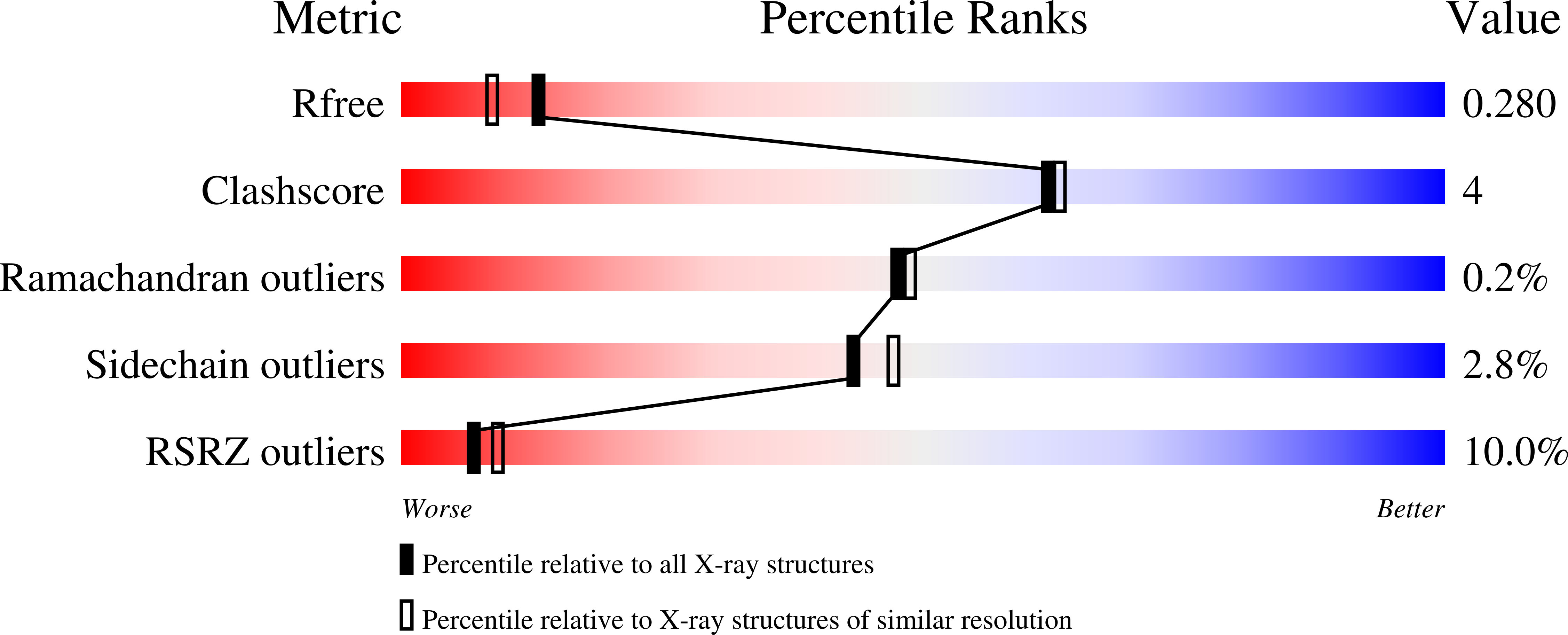
Ribosome Clearance by Fusb-Type Proteins Mediates Resistance to the Antibiotic Fusidic Acid
Cox, G., Thompson, G.S., Jenkins, H.T., Homans, S.W., Edwards, T.A., Oneill, A.J.(2012) Proc Natl Acad Sci U S A 109: 2102
- PubMed: 22308410
- DOI: https://doi.org/10.1073/pnas.1117275109
- Primary Citation of Related Structures:
2YB5 - PubMed Abstract:
Resistance to the antibiotic fusidic acid (FA) in the human pathogen Staphylococcus aureus usually results from expression of FusB-type proteins (FusB or FusC). These proteins bind to elongation factor G (EF-G), the target of FA, and rescue translation from FA-mediated inhibition by an unknown mechanism. Here we show that the FusB family are two-domain metalloproteins, the C-terminal domain of which contains a four-cysteine zinc finger with a unique structural fold. This domain mediates a high-affinity interaction with the C-terminal domains of EF-G. By binding to EF-G on the ribosome, FusB-type proteins promote the dissociation of stalled ribosome⋅EF-G⋅GDP complexes that form in the presence of FA, thereby allowing the ribosomes to resume translation. Ribosome clearance by these proteins represents a highly unusual antibiotic resistance mechanism, which appears to be fine-tuned by the relative abundance of FusB-type protein, ribosomes, and EF-G.
Organizational Affiliation:
Institute of Molecular and Cellular Biology, University of Leeds, Leeds LS2 9JT, United Kingdom.