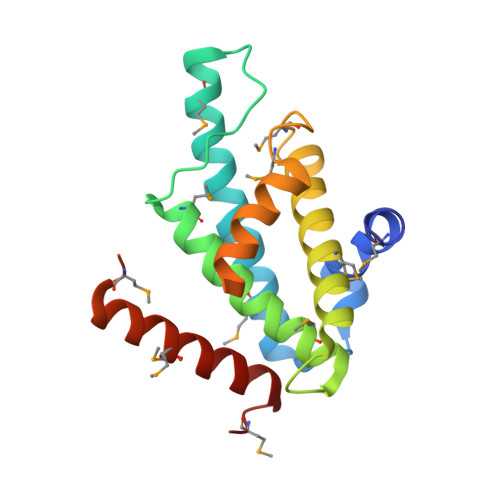
The Structural Basis for Recognition of Base J Containing DNA by a Novel DNA Binding Domain in Jbp1.
Heidebrecht, T., Christodoulou, E., Chalmers, M.J., Jan, S., Ter Riet, B., Grover, R.K., Joosten, R.P., Littler, D., Van Luenen, H., Griffin, P.R., Wentworth, P., Borst, P., Perrakis, A.(2011) Nucleic Acids Res 39: 5715
- PubMed: 21415010
- DOI: https://doi.org/10.1093/nar/gkr125
- Primary Citation of Related Structures:
2XSE - PubMed Abstract:
The J-binding protein 1 (JBP1) is essential for biosynthesis and maintenance of DNA base-J (β-d-glucosyl-hydroxymethyluracil). Base-J and JBP1 are confined to some pathogenic protozoa and are absent from higher eukaryotes, prokaryotes and viruses. We show that JBP1 recognizes J-containing DNA (J-DNA) through a 160-residue domain, DB-JBP1, with 10 000-fold preference over normal DNA. The crystal structure of DB-JBP1 revealed a helix-turn-helix variant fold, a 'helical bouquet' with a 'ribbon' helix encompassing the amino acids responsible for DNA binding. Mutation of a single residue (Asp525) in the ribbon helix abrogates specificity toward J-DNA. The same mutation renders JBP1 unable to rescue the targeted deletion of endogenous JBP1 genes in Leishmania and changes its distribution in the nucleus. Based on mutational analysis and hydrogen/deuterium-exchange mass-spectrometry data, a model of JBP1 bound to J-DNA was constructed and validated by small-angle X-ray scattering data. Our results open new possibilities for targeted prevention of J-DNA recognition as a therapeutic intervention for parasitic diseases.
Organizational Affiliation:
Division of Biochemistry, The Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam, The Netherlands.