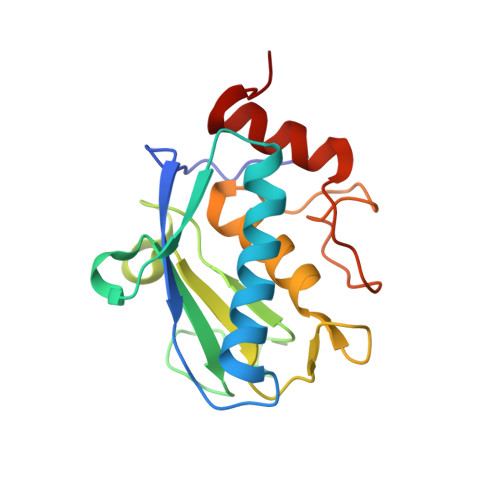
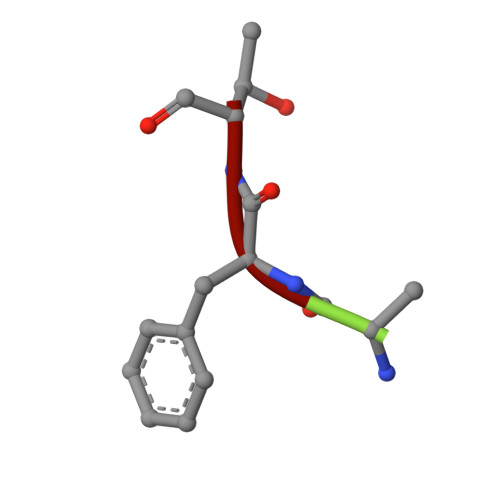
The Structure of the Catalytic Domain of Tannerella Forsythia Karilysin Reveals It is a Bacterial Xenologue of Animal Matrix Metalloproteinases.
Cerda-Costa, N., Guevara, T., Karim, A.Y., Ksiazek, M., Nguyen, K.A., Arolas, J.L., Potempa, J., Gomis-Ruth, F.X.(2011) Mol Microbiol 79: 119
- PubMed: 21166898
- DOI: https://doi.org/10.1111/j.1365-2958.2010.07434.x
- Primary Citation of Related Structures:
2XS3, 2XS4 - PubMed Abstract:
Metallopeptidases (MPs) are among virulence factors secreted by pathogenic bacteria at the site of infection. One such pathogen is Tannerella forsythia, a member of the microbial consortium that causes peridontitis, arguably the most prevalent infective chronic inflammatory disease known to mankind. The only reported MP secreted by T. forsythia is karilysin, a 52 kDa multidomain protein comprising a central 18 kDa catalytic domain (CD), termed Kly18, flanked by domains unrelated to any known protein. We analysed the 3D structure of Kly18 in the absence and presence of Mg(2+) or Ca(2+) , which are required for function and stability, and found that it evidences most of the structural features characteristic of the CDs of mammalian matrix metalloproteinases (MMPs). Unexpectedly, a peptide was bound to the active-site cleft of Kly18 mimicking a left-behind cleavage product, which revealed that the specificity pocket accommodates bulky hydrophobic side-chains of substrates as in mammalian MMPs. In addition, Kly18 displayed a unique Mg(2+) or Ca(2+) binding site and two flexible segments that could play a role in substrate binding. Phylogenetic and sequence similarity studies revealed that Kly18 is evolutionarily much closer to winged-insect and mammalian MMPs than to potential bacterial counterparts found by genomic sequencing projects. Therefore, we conclude that this first structurally characterized non-mammalian MMP is a xenologue co-opted through horizontal gene transfer during the intimate coexistence between T. forsythia and humans or other animals, in a very rare case of gene shuffling from eukaryotes to prokaryotes. Subsequently, this protein would have evolved in a bacterial environment to give rise to full-length karilysin that is furnished with unique flanking domains that do not conform to the general multidomain architecture of animal MMPs.
Organizational Affiliation:
Proteolysis Lab, Department of Structural Biology, Molecular Biology Institute of Barcelona, CSIC, Barcelona Science Park, Helix Building, c/ Baldiri Reixac, 15-21, E-08028 Barcelona, Catalunya, Spain.