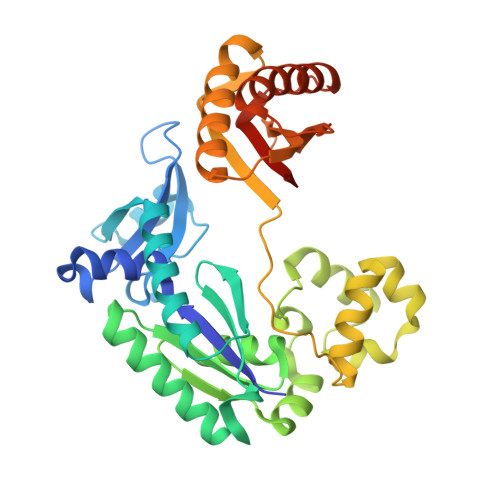
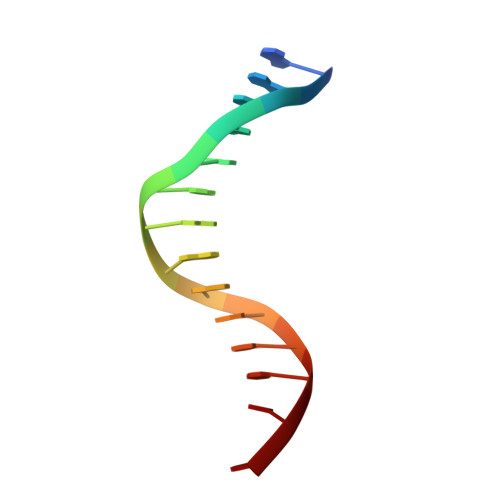
Metal Ion Dependence of the Active Site Conformation of the Trans-Lesion DNA Polymerase Dpo4 from Sulfolobus Solfataricus
Irimia, A., Loukachevitch, L.V., Eoff, R.L., Guengerich, F.P., Egli, M.(2010) Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 1013
- PubMed: 20823515
- DOI: https://doi.org/10.1107/S1744309110029374
- Primary Citation of Related Structures:
2XC9, 2XCA, 2XCP - PubMed Abstract:
Crystal structures of a binary Mg2+-form Dpo4-DNA complex with 1,N2-etheno-dG in the template strand as well as of ternary Mg2+-form Dpo4-DNA-dCTP/dGTP complexes with 8-oxoG in the template strand have been determined. Comparison of their conformations and active-site geometries with those of the corresponding Ca2+-form complexes revealed that the DNA and polymerase undergo subtle changes as a result of the catalytically more active Mg2+ occupying both the A and B sites.
Organizational Affiliation:
Department of Biochemistry and Center in Molecular Toxicology, Vanderbilt University, Nashville, TN 37232, USA.