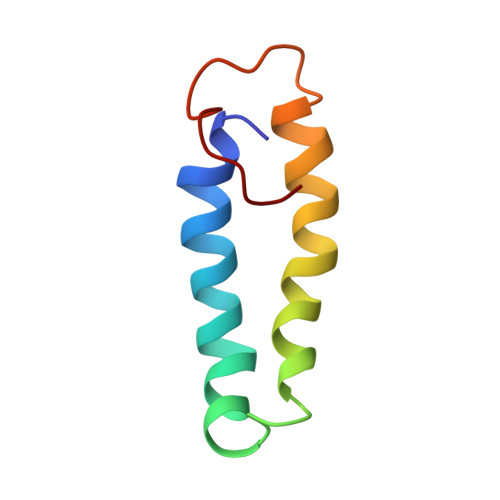
Structural Basis of Moelcular Recognition by the Leishmania Small Hydrophilic Endoplasmic Reticulum-Associated Protein, Sherp, at Membrane Surfaces
Moore, B., Miles, A.J., Guerra, C.G., Simpson, P., Iwata, M., Wallace, B.A., Matthews, S.J., Smith, D.F., Brown, K.A.(2011) J Biol Chem 286: 9246
- PubMed: 21106528
- DOI: https://doi.org/10.1074/jbc.M110.130427
- Primary Citation of Related Structures:
2X43 - PubMed Abstract:
The 57-residue small hydrophilic endoplasmic reticulum-associated protein (SHERP) shows highly specific, stage-regulated expression in the non-replicative vector-transmitted stages of the kinetoplastid parasite, Leishmania major, the causative agent of human cutaneous leishmaniasis. Previous studies have demonstrated that SHERP localizes as a peripheral membrane protein on the cytosolic face of the endoplasmic reticulum and on outer mitochondrial membranes, whereas its high copy number suggests a critical function in vivo. However, the absence of defined domains or identifiable orthologues, together with lack of a clear phenotype in transgenic parasites lacking SHERP, has limited functional understanding of this protein. Here, we use a combination of biophysical and biochemical methods to demonstrate that SHERP can be induced to adopt a globular fold in the presence of anionic lipids or SDS. Cross-linking and binding studies suggest that SHERP has the potential to form a complex with the vacuolar type H(+)-ATPase. Taken together, these results suggest that SHERP may function in modulating cellular processes related to membrane organization and/or acidification during vector transmission of infective Leishmania.
Organizational Affiliation:
Division of Cell and Molecular Biology, Centre for Molecular Microbiology and Infection, Imperial College London, Exhibition Road, London SW7 2AZ, United Kingdom.